Working with Individual Scans: H5ParcellatedScan and H5ParcellatedScanSummary
fmristore Package
2025-08-19
Source:vignettes/H5ParcellatedScan.Rmd
H5ParcellatedScan.Rmd
library(fmristore)
#>
#> Attaching package: 'fmristore'
#> The following object is masked from 'package:stats':
#>
#> offset
library(neuroim2)
#> Loading required package: Matrix
#>
#> Attaching package: 'neuroim2'
#> The following object is masked from 'package:base':
#>
#> scaleOverview: The Individual Scan Components
In the fmristore parcellated data system, H5ParcellatedMultiScan serves as a container that holds multiple individual scan objects. Each scan within the container is either:
- H5ParcellatedScan: Full voxel-level time series data
- H5ParcellatedScanSummary: Cluster-averaged time series data
This vignette focuses on working with these individual scan objects - how to access them from the parent H5ParcellatedMultiScan container and what operations you can perform on each scan type.
Step 1: Create Sample Data Using the New Interface
First, let’s create a sample dataset using the modern
write_dataset() and read_dataset()
functions:
# Create a small brain with realistic structure
brain_dims <- c(10, 10, 5)
brain_space <- NeuroSpace(brain_dims, spacing = c(2, 2, 2))
# Define brain mask (central region)
mask_data <- array(FALSE, brain_dims)
mask_data[4:7, 4:7, 2:4] <- TRUE # Small brain region
mask <- LogicalNeuroVol(mask_data, brain_space)
# Divide brain into 3 clusters (regions)
n_voxels <- sum(mask)
cluster_assignments <- rep(1:3, length.out = n_voxels)
clusters <- ClusteredNeuroVol(mask, cluster_assignments)
cat("Created brain with", n_voxels, "voxels in", max(cluster_assignments), "clusters\n")
#> Created brain with 48 voxels in 3 clusters
print(table(clusters@clusters))
#>
#> 1 2 3
#> 16 16 16
Create Multiple fMRI Runs with Different Characteristics
# Create 3 different fMRI runs
run_names <- c("task_full", "rest_full", "task_summary")
runs_data <- list()
# Run 1: Task run (full data) - 60 timepoints
n_time1 <- 60
fmri_dims1 <- c(brain_dims, n_time1)
fmri_data1 <- array(rnorm(prod(fmri_dims1)), dim = fmri_dims1)
# Add task activation to cluster 2 (motor region)
cluster_2_voxels <- which(clusters@clusters == 2)
mask_indices <- which(mask, arr.ind = TRUE)
for (v in cluster_2_voxels) {
x <- mask_indices[v, 1]
y <- mask_indices[v, 2]
z <- mask_indices[v, 3]
# Add sinusoidal activation
fmri_data1[x, y, z, ] <- fmri_data1[x, y, z, ] + 0.8 * sin(seq(0, 4 * pi, length.out = n_time1))
}
runs_data[[1]] <- NeuroVec(fmri_data1, NeuroSpace(fmri_dims1))
# Run 2: Rest run (full data) - 50 timepoints
n_time2 <- 50
fmri_dims2 <- c(brain_dims, n_time2)
fmri_data2 <- array(rnorm(prod(fmri_dims2)), dim = fmri_dims2)
# Add resting state connectivity pattern
for (i in 1:3) {
cluster_voxels <- which(clusters@clusters == i)
shared_signal <- rnorm(n_time2, sd = 0.3)
for (v in cluster_voxels) {
x <- mask_indices[v, 1]
y <- mask_indices[v, 2]
z <- mask_indices[v, 3]
fmri_data2[x, y, z, ] <- fmri_data2[x, y, z, ] + shared_signal
}
}
runs_data[[2]] <- NeuroVec(fmri_data2, NeuroSpace(fmri_dims2))
# Run 3: Will be saved as summary data (we'll use run 1's data)
runs_data[[3]] <- runs_data[[1]]
cat(
"Created", length(runs_data), "runs with timepoints:",
paste(sapply(runs_data, function(x) dim(x)[4]), collapse = ", "), "\n"
)
#> Created 3 runs with timepoints: 60, 50, 60Save Mixed Full and Summary Data
# Save to HDF5 file - mix of full and summary scans
h5_file <- tempfile(fileext = ".h5")
# First, save the two full runs
write_dataset(list(runs_data[[1]], runs_data[[2]]),
file = h5_file,
clusters = clusters,
scan_names = c("task_full", "rest_full"),
summary = FALSE
) # Full voxel data
#> Creating HDF5 file: /tmp/RtmpasnhTW/file2318413d5c87.h5
#> Writing global structures (mask, clusters, header)...
#> Writing scan data...
#> Writing scan: task_full (type: full)
#> Scan 'task_full': nTime not in metadata, attempting to infer from sdata list.
#> Scan 'task_full': Inferred nTime 60 from sdata item 'cluster_1'.
#> Writing scan: rest_full (type: full)
#> Scan 'rest_full': nTime not in metadata, attempting to infer from sdata list.
#> Scan 'rest_full': Inferred nTime 50 from sdata item 'cluster_1'.
# Then add a summary run to the same file
# Note: In practice, you'd append this or use write_clustered_experiment_h5()
# For this example, we'll create separate files to show both types clearly
summary_file <- tempfile(fileext = ".h5")
write_dataset(runs_data[[3]],
file = summary_file,
clusters = clusters,
scan_names = "task_summary",
summary = TRUE
) # Cluster averages only
#> Creating HDF5 file: /tmp/RtmpasnhTW/file23181c1ab41e.h5
#> Writing global structures (mask, clusters, header)...
#> Writing scan data...
#> Writing scan: scan_001 (type: summary)
cat("Created files:\n")
#> Created files:
cat(" Full data file:", round(file.size(h5_file) / 1024, 1), "KB\n")
#> Full data file: 55 KB
cat(" Summary data file:", round(file.size(summary_file) / 1024, 1), "KB\n")
#> Summary data file: 20.1 KBStep 2: Load the Data with read_dataset()
# Load the datasets using the new read_dataset() function
full_experiment <- read_dataset(h5_file)
#> Detected type 'H5ParcellatedMultiScan' in file: /tmp/RtmpasnhTW/file2318413d5c87.h5
#> Clusters argument is NULL, attempting to read from HDF5 file (/cluster_map).
summary_experiment <- read_dataset(summary_file)
#> Detected type 'H5ParcellatedMultiScan' in file: /tmp/RtmpasnhTW/file23181c1ab41e.h5
#> Clusters argument is NULL, attempting to read from HDF5 file (/cluster_map).
cat("Full data experiment:\n")
#> Full data experiment:
print(full_experiment)
#>
#> H5ParcellatedMultiScan
#> # runs : 2
#> # clusters : 0
#> mask dims : 10x10x5
#> HDF5 file : /tmp/RtmpasnhTW/file2318413d5c87.h5
cat("\nSummary data experiment:\n")
#>
#> Summary data experiment:
print(summary_experiment)
#>
#> H5ParcellatedMultiScan
#> # runs : 1
#> # clusters : 0
#> mask dims : 10x10x5
#> HDF5 file : /tmp/RtmpasnhTW/file23181c1ab41e.h5Step 3: Accessing Individual Scans from the Container
This is the key concept: H5ParcellatedScan and H5ParcellatedScanSummary objects are accessed from their parent H5ParcellatedMultiScan container.
# Method 1: Access by position in @runs slot
task_scan <- full_experiment@runs[[1]] # First scan
rest_scan <- full_experiment@runs[[2]] # Second scan
# Method 2: Access by name if names are available
scan_names_full <- scan_names(full_experiment)
cat("Available scan names in full experiment:", paste(scan_names_full, collapse = ", "), "\n")
#> Available scan names in full experiment: rest_full, task_full
if (length(scan_names_full) > 0) {
task_scan_by_name <- full_experiment@runs[[scan_names_full[1]]]
cat("Accessed scan by name:", scan_names_full[1], "\n")
}
#> Accessed scan by name: rest_full
# Show what type of scan objects we got
cat("Task scan class:", class(task_scan), "\n")
#> Task scan class: H5ParcellatedScan
cat("Rest scan class:", class(rest_scan), "\n")
#> Rest scan class: H5ParcellatedScan
# Access the summary scan
summary_scan <- summary_experiment@runs[[1]]
cat("Summary scan class:", class(summary_scan), "\n")
#> Summary scan class: H5ParcellatedScanSummaryUnderstanding Individual Scan Properties
# Each individual scan has its own properties
cat("=== Task Scan (H5ParcellatedScan) ===\n")
#> === Task Scan (H5ParcellatedScan) ===
cat("Dimensions:", paste(dim(task_scan), collapse = " × "), "\n")
#> Dimensions: 10 × 10 × 5 × 50
cat("Class:", class(task_scan), "\n")
#> Class: H5ParcellatedScan
cat("Number of voxels:", dim(task_scan)[1], "\n")
#> Number of voxels: 10
cat("Number of timepoints:", dim(task_scan)[2], "\n\n")
#> Number of timepoints: 10
cat("=== Summary Scan (H5ParcellatedScanSummary) ===\n")
#> === Summary Scan (H5ParcellatedScanSummary) ===
summary_matrix <- as.matrix(summary_scan)
cat("Summary dimensions:", paste(dim(summary_matrix), collapse = " × "), "\n")
#> Summary dimensions: 60 × 3
cat("Class:", class(summary_scan), "\n")
#> Class: H5ParcellatedScanSummary
cat("Number of clusters:", ncol(summary_matrix), "\n")
#> Number of clusters: 3
cat("Number of timepoints:", nrow(summary_matrix), "\n\n")
#> Number of timepoints: 60
cat("=== Storage Efficiency ===\n")
#> === Storage Efficiency ===
n_voxels_full <- dim(task_scan)[1] * dim(task_scan)[2]
n_elements_summary <- prod(dim(summary_matrix))
cat("Full scan elements:", n_voxels_full, "\n")
#> Full scan elements: 100
cat("Summary scan elements:", n_elements_summary, "\n")
#> Summary scan elements: 180
cat("Compression ratio:", round(n_voxels_full / n_elements_summary, 1), ":1\n")
#> Compression ratio: 0.6 :1H5ParcellatedScan: Operations on Individual Full Scans
When working with H5ParcellatedScan objects (full voxel data), you can extract time series for specific voxels and perform voxel-level analyses.
Extract Time Series from Individual Voxels
# Extract time series for specific voxel indices
voxel_indices <- c(1, 5, 10, 15)
task_voxel_ts <- series(task_scan, i = voxel_indices)
# Show what we extracted
cat("Extracted time series dimensions:", paste(dim(task_voxel_ts), collapse = " × "), "\n")
#> Extracted time series dimensions: 50 × 4
cat("(rows = timepoints, columns = voxels)\n\n")
#> (rows = timepoints, columns = voxels)
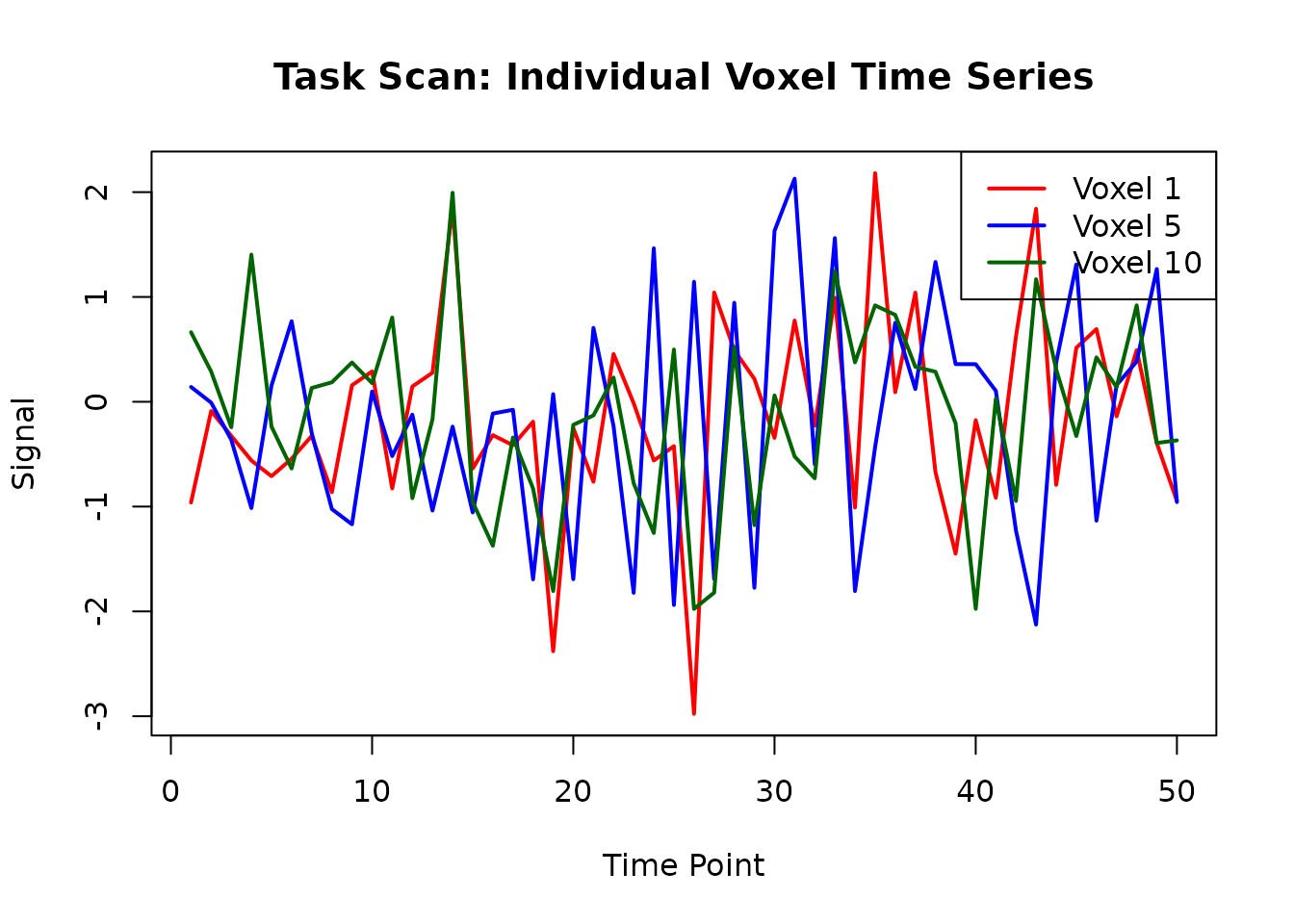
# Plot individual voxel time series
matplot(task_voxel_ts[, 1:3],
type = "l", lty = 1, lwd = 2,
main = "Task Scan: Individual Voxel Time Series",
xlab = "Time Point", ylab = "Signal",
col = c("red", "blue", "darkgreen")
)
legend("topright",
legend = paste("Voxel", voxel_indices[1:3]),
col = c("red", "blue", "darkgreen"), lty = 1, lwd = 2
)
Extract Time Series for All Voxels in a Cluster
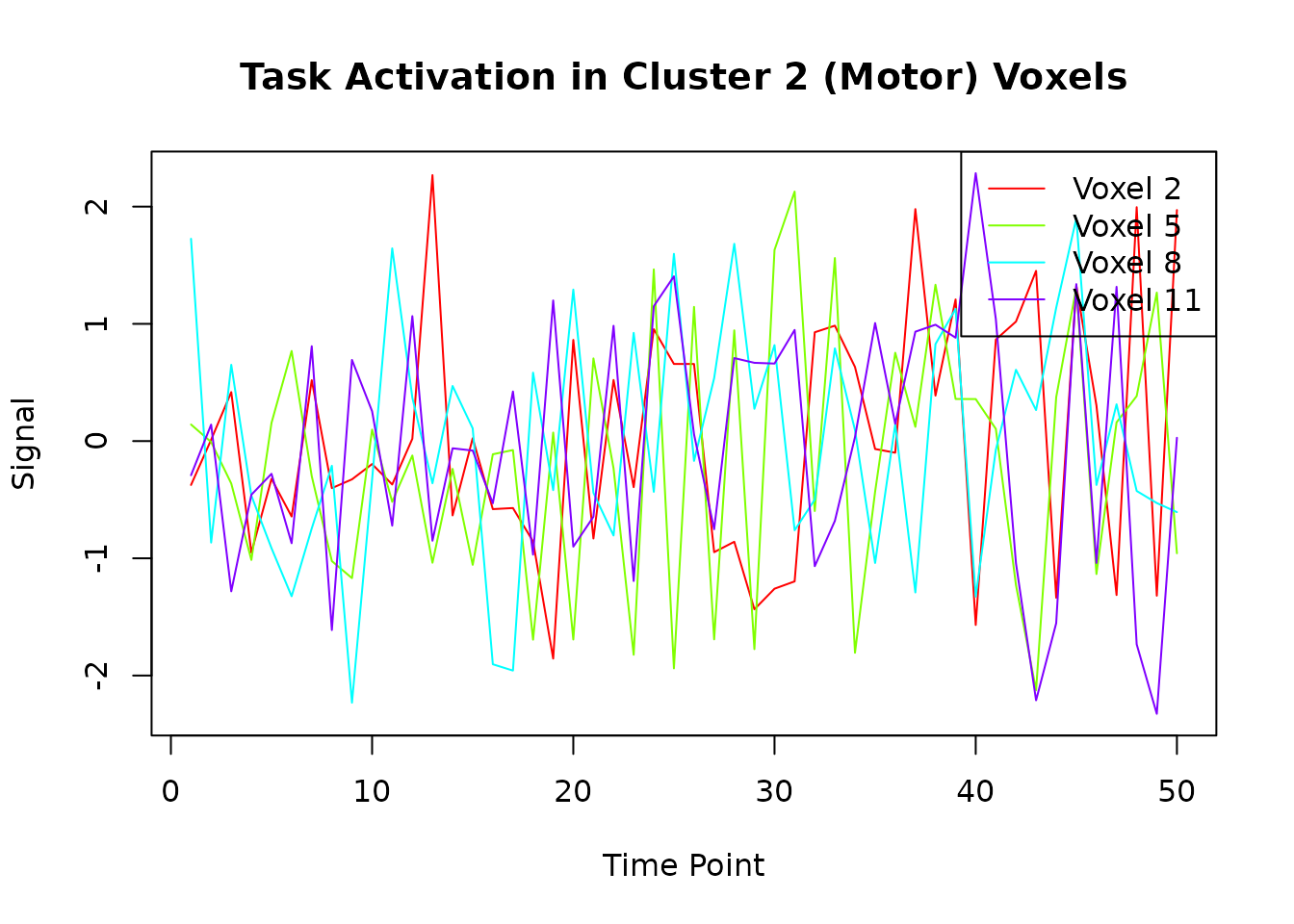
# Find all voxels in cluster 2 (the motor region with task activation)
cluster_2_voxels <- which(clusters@clusters == 2)
cat("Cluster 2 contains", length(cluster_2_voxels), "voxels\n")
#> Cluster 2 contains 16 voxels
# Extract time series for all voxels in cluster 2
cluster_2_ts <- series(task_scan, i = cluster_2_voxels)
cat("Cluster 2 time series dimensions:", paste(dim(cluster_2_ts), collapse = " × "), "\n")
#> Cluster 2 time series dimensions: 50 × 16
# Plot the first few voxels from cluster 2 to see the task activation
matplot(cluster_2_ts[, 1:min(4, ncol(cluster_2_ts))],
type = "l", lty = 1,
main = "Task Activation in Cluster 2 (Motor) Voxels",
xlab = "Time Point", ylab = "Signal",
col = rainbow(min(4, ncol(cluster_2_ts)))
)
legend("topright",
legend = paste("Voxel", cluster_2_voxels[1:min(4, ncol(cluster_2_ts))]),
col = rainbow(min(4, ncol(cluster_2_ts))), lty = 1
)
Compare Task vs Rest Scans (Individual Scan Analysis)
# Extract the same voxel indices from both scans
task_sample <- series(task_scan, i = 1:6)
rest_sample <- series(rest_scan, i = 1:6)
# Calculate variance for each voxel in each scan
task_vars <- apply(task_sample, 2, var)
rest_vars <- apply(rest_sample, 2, var)
# Compare
comparison_df <- data.frame(
Voxel = 1:6,
Task_Variance = task_vars,
Rest_Variance = rest_vars,
Ratio = task_vars / rest_vars
)
print("Variance comparison between task and rest scans:")
#> [1] "Variance comparison between task and rest scans:"
print(round(comparison_df, 3))
#> Voxel Task_Variance Rest_Variance Ratio
#> 1 1 0.874 1.048 0.834
#> 2 2 1.067 1.559 0.684
#> 3 3 1.459 0.771 1.892
#> 4 4 0.801 0.729 1.099
#> 5 5 1.163 1.294 0.899
#> 6 6 1.050 1.095 0.958
# Plot comparison
par(mfrow = c(1, 2))
plot(task_vars, rest_vars,
xlab = "Task Variance", ylab = "Rest Variance",
main = "Variance Comparison", pch = 19, col = "blue"
)
abline(0, 1, col = "red", lty = 2)
plot(1:6, comparison_df$Ratio,
type = "b", pch = 19, col = "darkgreen",
xlab = "Voxel Index", ylab = "Task/Rest Variance Ratio",
main = "Variance Ratio (Task/Rest)"
)
abline(h = 1, col = "red", lty = 2)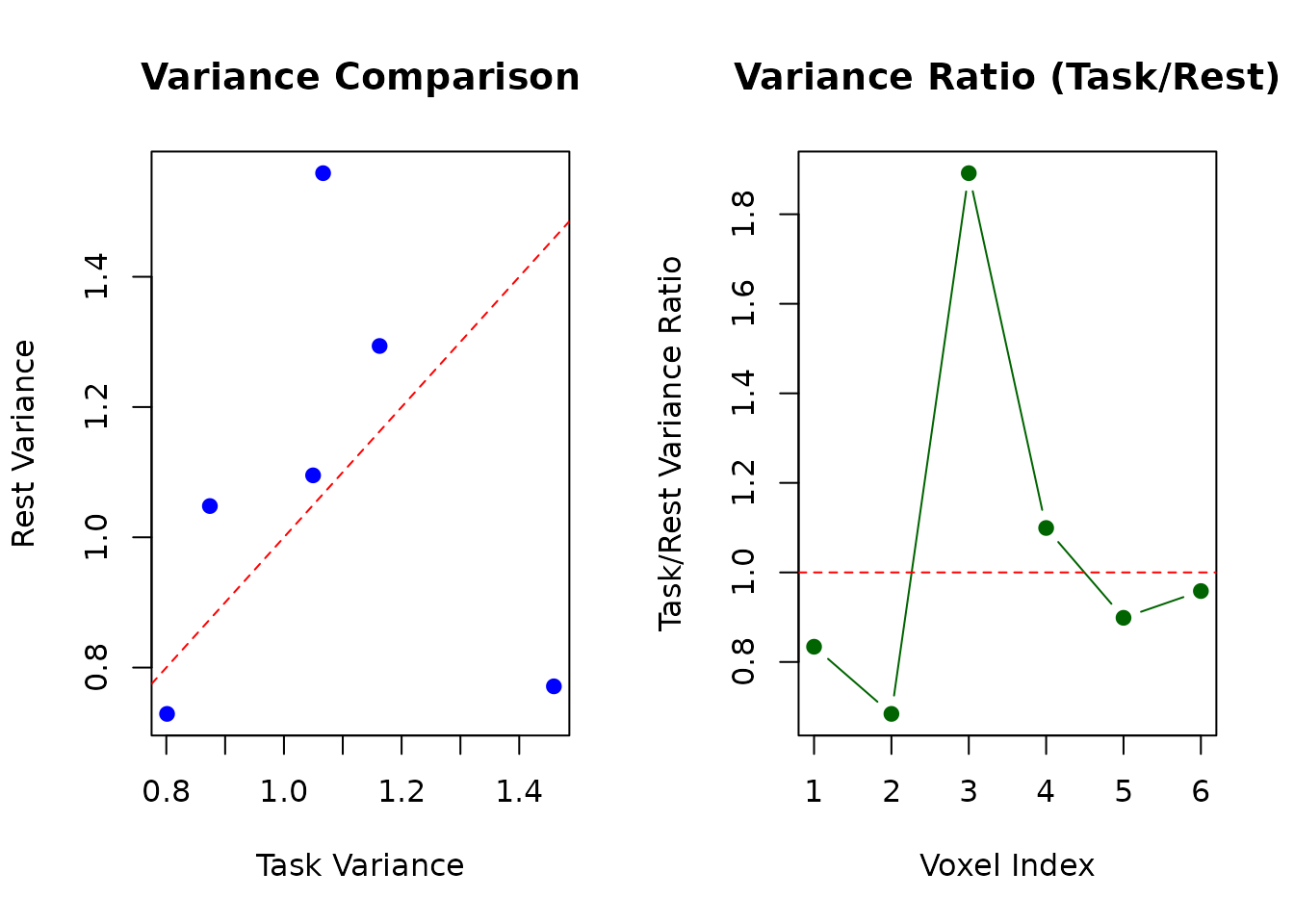
H5ParcellatedScanSummary: Operations on Individual Summary Scans
When working with H5ParcellatedScanSummary objects, you work with cluster-averaged data that’s much more compact.
Extract and Analyze Summary Data
# Get the summary matrix (time × clusters)
summary_matrix <- as.matrix(summary_scan)
cat("Summary matrix dimensions:", paste(dim(summary_matrix), collapse = " × "), "\n")
#> Summary matrix dimensions: 60 × 3
cat("Timepoints:", nrow(summary_matrix), ", Clusters:", ncol(summary_matrix), "\n\n")
#> Timepoints: 60 , Clusters: 3
# Show column names if available
if (!is.null(colnames(summary_matrix))) {
cat("Cluster names:", paste(colnames(summary_matrix), collapse = ", "), "\n")
} else {
cat("Using default cluster numbering: 1 to", ncol(summary_matrix), "\n")
}
#> Cluster names: Cluster1, Cluster2, Cluster3
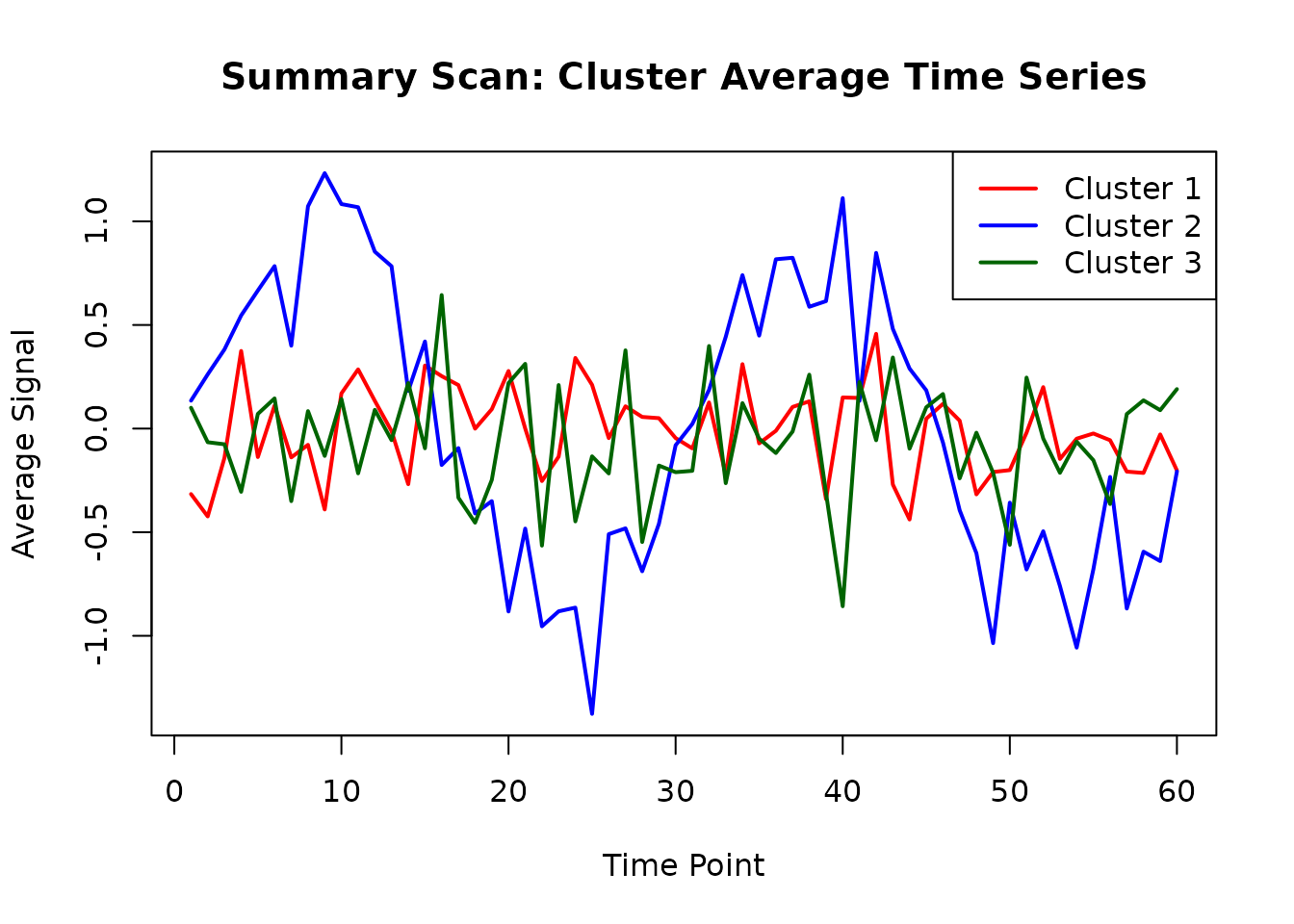
# Plot cluster averages over time
matplot(summary_matrix,
type = "l", lty = 1, lwd = 2,
main = "Summary Scan: Cluster Average Time Series",
xlab = "Time Point", ylab = "Average Signal",
col = c("red", "blue", "darkgreen")
)
legend("topright",
legend = paste("Cluster", 1:ncol(summary_matrix)),
col = c("red", "blue", "darkgreen"), lty = 1, lwd = 2
)
Compute Cluster-to-Cluster Correlations
# Calculate correlation matrix between clusters
cluster_correlations <- cor(summary_matrix)
cat("Cluster correlation matrix:\n")
#> Cluster correlation matrix:
print(round(cluster_correlations, 3))
#> Cluster1 Cluster2 Cluster3
#> Cluster1 1.000 0.086 0.059
#> Cluster2 0.086 1.000 0.047
#> Cluster3 0.059 0.047 1.000
# Visualize correlation matrix
if (requireNamespace("corrplot", quietly = TRUE)) {
corrplot::corrplot(cluster_correlations,
method = "color",
title = "Inter-Cluster Correlations",
mar = c(0, 0, 2, 0)
)
} else {
# Simple heatmap alternative
image(1:ncol(cluster_correlations), 1:nrow(cluster_correlations),
as.matrix(cluster_correlations),
main = "Inter-Cluster Correlations",
xlab = "Cluster", ylab = "Cluster",
col = heat.colors(20)
)
}
Practical Usage Guidelines
Choose H5ParcellatedScan when you need:
Individual voxel analysis within clusters:
# Example: Find the most variable voxel in each cluster
most_variable_voxels <- sapply(1:3, function(cluster_id) {
cluster_voxels <- which(clusters@clusters == cluster_id)
cluster_ts <- series(task_scan, i = cluster_voxels)
variances <- apply(cluster_ts, 2, var)
cluster_voxels[which.max(variances)]
})Voxel-wise statistical analyses:
# Example: Correlate each voxel with a task regressor
task_regressor <- sin(seq(0, 4 * pi, length.out = dim(task_scan)[2]))
all_voxel_ts <- series(task_scan, i = 1:dim(task_scan)[1])
voxel_correlations <- cor(all_voxel_ts, task_regressor)Spatial pattern analysis within regions:
Choose H5ParcellatedScanSummary when you need:
Network connectivity analysis:
# Already computed above - cluster correlation matrix
print("Inter-cluster connectivity strengths:")
#> [1] "Inter-cluster connectivity strengths:"
diag(cluster_correlations) <- NA # Remove self-correlations
print(round(cluster_correlations, 3))
#> Cluster1 Cluster2 Cluster3
#> Cluster1 NA 0.086 0.059
#> Cluster2 0.086 NA 0.047
#> Cluster3 0.059 0.047 NAEfficient group comparisons:
# Example: Compare average activation levels between conditions
# If you had multiple summary scans, you could do:
cluster_means <- colMeans(summary_matrix)
names(cluster_means) <- paste("Cluster", 1:length(cluster_means))
print("Average activation per cluster:")
#> [1] "Average activation per cluster:"
print(round(cluster_means, 3))
#> Cluster 1 Cluster 2 Cluster 3
#> -0.012 0.004 -0.058Memory-efficient temporal analysis:
# Example: Analyze temporal dynamics at cluster level
cluster_peak_times <- apply(summary_matrix, 2, which.max)
names(cluster_peak_times) <- paste("Cluster", 1:length(cluster_peak_times))
print("Peak activation timepoints per cluster:")
#> [1] "Peak activation timepoints per cluster:"
print(cluster_peak_times)
#> Cluster 1 Cluster 2 Cluster 3
#> 42 9 16Working with Both Scan Types in Practice
When you have an H5ParcellatedMultiScan container with mixed scan types:
# Access different scan types from the same experiment
full_scan1 <- full_experiment@runs[["task_full"]] # H5ParcellatedScan
full_scan2 <- full_experiment@runs[["rest_full"]] # H5ParcellatedScan
summary_scan <- summary_experiment@runs[["task_summary"]] # H5ParcellatedScanSummary
# Use appropriate methods for each type
voxel_data <- series(full_scan1, i = 1:10) # Voxel time series
cluster_data <- as.matrix(summary_scan) # Cluster averages
# Cannot use series() on summary scans - this would error:
# bad_call <- series(summary_scan, i = 1:5) # ERROR!Key Individual Scan Operations Summary
| Operation | H5ParcellatedScan | H5ParcellatedScanSummary |
|---|---|---|
series(scan, i = indices) |
✅ Extract voxel time series | ❌ Not available |
as.matrix(scan) |
❌ Not available | ✅ Get cluster × time matrix |
dim(scan) |
✅ [voxels, timepoints] | ❌ Use dim(as.matrix(scan))
|
scan[i, j, k, t] |
✅ 4D indexing | ❌ Not available |
| Memory efficiency | Lower (full voxel data) | Higher (cluster averages only) |
Memory and Performance Considerations
# Compare memory footprint of different scan types
cat("Memory usage comparison:\n")
#> Memory usage comparison:
cat(" Task scan object size:", format(object.size(task_scan), units = "KB"), "\n")
#> Task scan object size: 28.4 Kb
cat(" Summary scan object size:", format(object.size(summary_scan), units = "KB"), "\n")
#> Summary scan object size: 29 Kb
cat(" Summary matrix size:", format(object.size(summary_matrix), units = "KB"), "\n\n")
#> Summary matrix size: 2.1 Kb
# File size comparison
cat("File size comparison:\n")
#> File size comparison:
cat(" Full data file:", round(file.size(h5_file) / 1024, 1), "KB\n")
#> Full data file: 55 KB
cat(" Summary data file:", round(file.size(summary_file) / 1024, 1), "KB\n")
#> Summary data file: 20.1 KBKey Takeaways
Individual scan objects (
H5ParcellatedScanandH5ParcellatedScanSummary) are accessed from H5ParcellatedMultiScan containers via@runs[[index]]or@runs[["name"]]-
H5ParcellatedScan provides voxel-level access within clusters:
- Use
series(scan, i = indices)for individual voxel time series - Supports 4D indexing:
scan[i, j, k, t] - Essential for spatial pattern analysis and voxel-wise statistics
- Use
-
H5ParcellatedScanSummary provides efficient cluster-level analysis:
- Use
as.matrix(scan)to get time × cluster matrix - Perfect for connectivity analysis and group comparisons
- Much smaller memory footprint
- Use
Modern workflow uses
read_dataset()andwrite_dataset()functions for simplified data I/O with automatic type detection-
Choose the right scan type based on your analysis needs:
- Full scans for detailed within-cluster analysis
- Summary scans for between-cluster connectivity and efficiency
Next Steps
-
For container management: See
vignette("H5ParcellatedMultiScan")for working with multiple runs -
For data creation: Learn about the new
write_dataset()interface for saving parcellated data - For advanced analysis: Explore mixed designs with both full and summary scans in the same container