Event Models
Bradley R. Buchsbaum
2025-02-22
Source:vignettes/a_04_event_models.Rmd
a_04_event_models.RmdfMRI event models
An “event model” is a description of an fMRI experiment, which consists of a collection of experimental “events” (stimuli/conditions/responses) which are embedded in an experimental design. By convolving each event type with a hemodynamic response function we from a set of regressors that are used to model and make inferences about the BOLD response.
A simple fMRI design
Consider a basic design, consisting of four stimulus types, each repeated 4 times over the course of a single scanning run. The stimuli are pictures of faces, scenes, tools and object presented for 2s a piece in a pseudorandom order. There is a uniformly distributed (4-7s) inter-stimulus interval (ISI) separating each stimulus presentation.
##
## Attaching package: 'fmrireg'## The following object is masked from 'package:stats':
##
## convolveNow we contruct a “design table” by randomly sampling from the four conditions:
simple_design <- data.frame(stim=factor(sample(rep(cond, 4))),
ISI=sample(4:7, NSTIM, replace=TRUE),
run=rep(1, NSTIM),
trial=factor(1:NSTIM))Event onsets are calculated as the cumulative sum of the ISI variables. Then we construct a sampling_frame which define the temporal sampling and block structure of the experiment. Here we have only 1 block and a repetition frequency (TR) of 2.
simple_design$onset <- cumsum(simple_design$ISI+2) -2
TR <- 2
sframe <- sampling_frame(blocklens=70, TR)Now the fun part. We create an event_model that describes the experimental model in terms of the stimulus onsets, conditions, and hemodynamic response function (‘hrf’).
emodel <- event_model(onset ~ fmrireg::hrf(stim, basis=fmrireg::HRF_SPMG1), data=simple_design, block = ~ run, sampling_frame=sframe)
print(emodel)##
## ╔══════════════════════════════════════════╗
## ║ fMRI Event Model ║
## ╚══════════════════════════════════════════╝
##
## Model Formula:
## onset ~ fmrireg::hrf(stim, basis = fmrireg::HRF_SPMG1)
##
## 📈 Model Summary:
## • Number of Terms: 1
## • Total Events: 16
## • Design Matrix Columns: 4
## • Number of Blocks: 1
##
## 📋 Event Table Preview:
## • Variables: stim, ISI, run, trial, onset
## stim ISI run trial onset
## 1 face 6 1 1 6
## 2 tool 4 1 2 12
## 3 object 4 1 3 18
## ... ( 13 more rows )
##
## 🔍 Model Terms:
##
## Term 1 : stim
## • Conditions: stim[face], stim[object], stim[scene], stim[tool]
## • HRF Type: SPMG1
## • Basis Functions: 1And we can plot the model:
plot(emodel)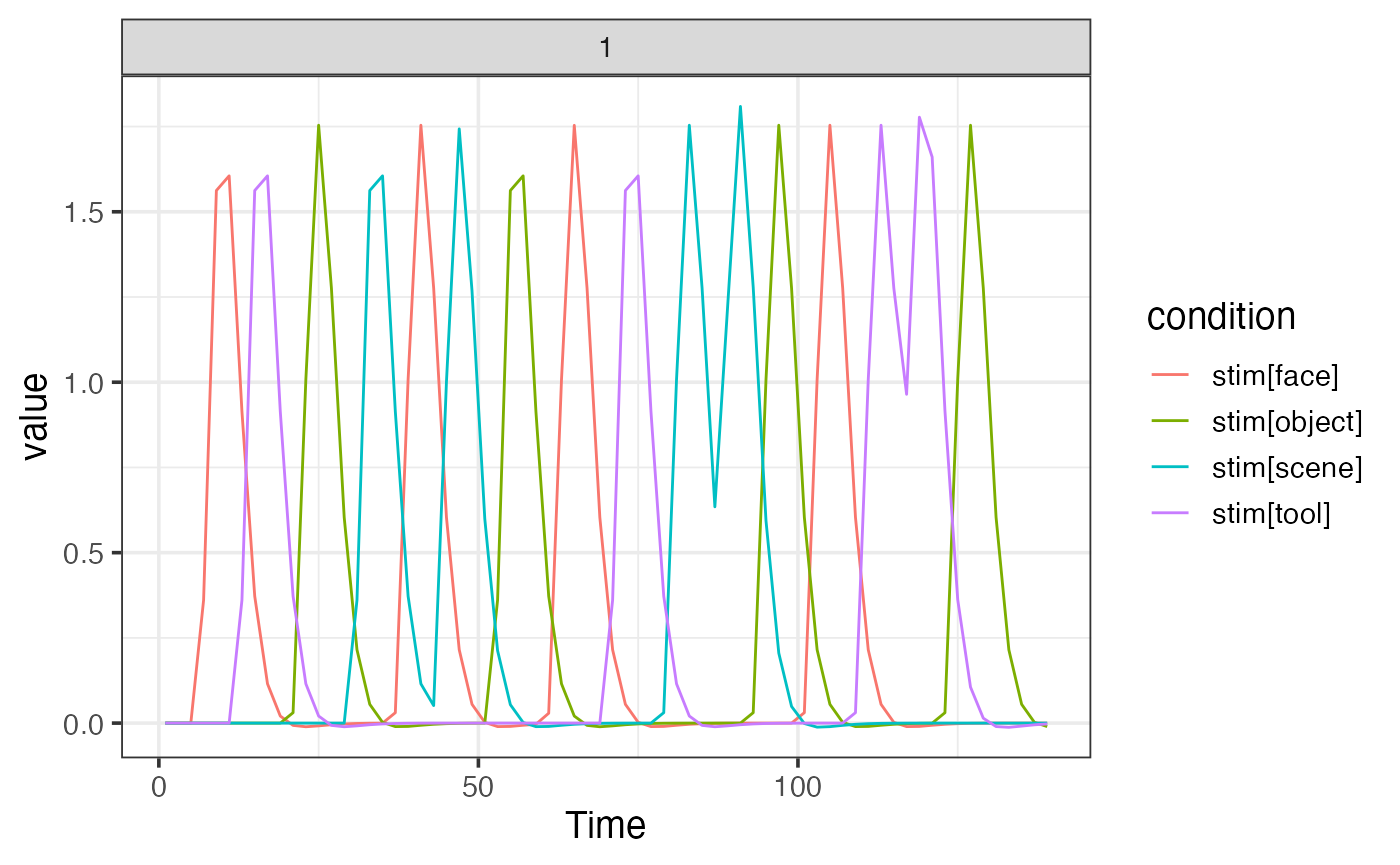
A simple fMRI design with multiple blocks
cond <- c("face", "scene", "tool", "object")
NSTIM <- length(cond)*4
## construct a design table with two blocks
design1 <- data.frame(stim=factor(sample(rep(cond, 4))),
ISI=sample(4:7, 16, replace=TRUE),
run=rep(1, NSTIM))
design2 <- data.frame(stim=factor(sample(rep(cond, 4))),
ISI=sample(4:7, 16, replace=TRUE),
run=rep(2, NSTIM))
design1$onset <- cumsum(design1$ISI+2) -2
design2$onset <- cumsum(design2$ISI+2) -2
design <- rbind(design1, design2)
## create a 'sampling_frame' describing the temporal outline of the scan consisting of 70 images collected every once every 2 seconds.
TR <- 2
sframe <- sampling_frame(blocklens=c(70,70), TR)
## create the event model d
emodel <- event_model(onset ~ hrf(stim), data=design, block = ~ run, sampling_frame=sframe)
print(emodel)##
## ╔══════════════════════════════════════════╗
## ║ fMRI Event Model ║
## ╚══════════════════════════════════════════╝
##
## Model Formula:
## onset ~ hrf(stim)
##
## 📈 Model Summary:
## • Number of Terms: 1
## • Total Events: 32
## • Design Matrix Columns: 4
## • Number of Blocks: 2
##
## 📋 Event Table Preview:
## • Variables: stim, ISI, run, onset
## stim ISI run onset
## 1 tool 4 1 4
## 2 face 4 1 10
## 3 scene 6 1 18
## ... ( 29 more rows )
##
## 🔍 Model Terms:
##
## Term 1 : stim
## • Conditions: stim[face], stim[object], stim[scene], stim[tool]
## • HRF Type: spmg1
## • Basis Functions: 1
plot(emodel)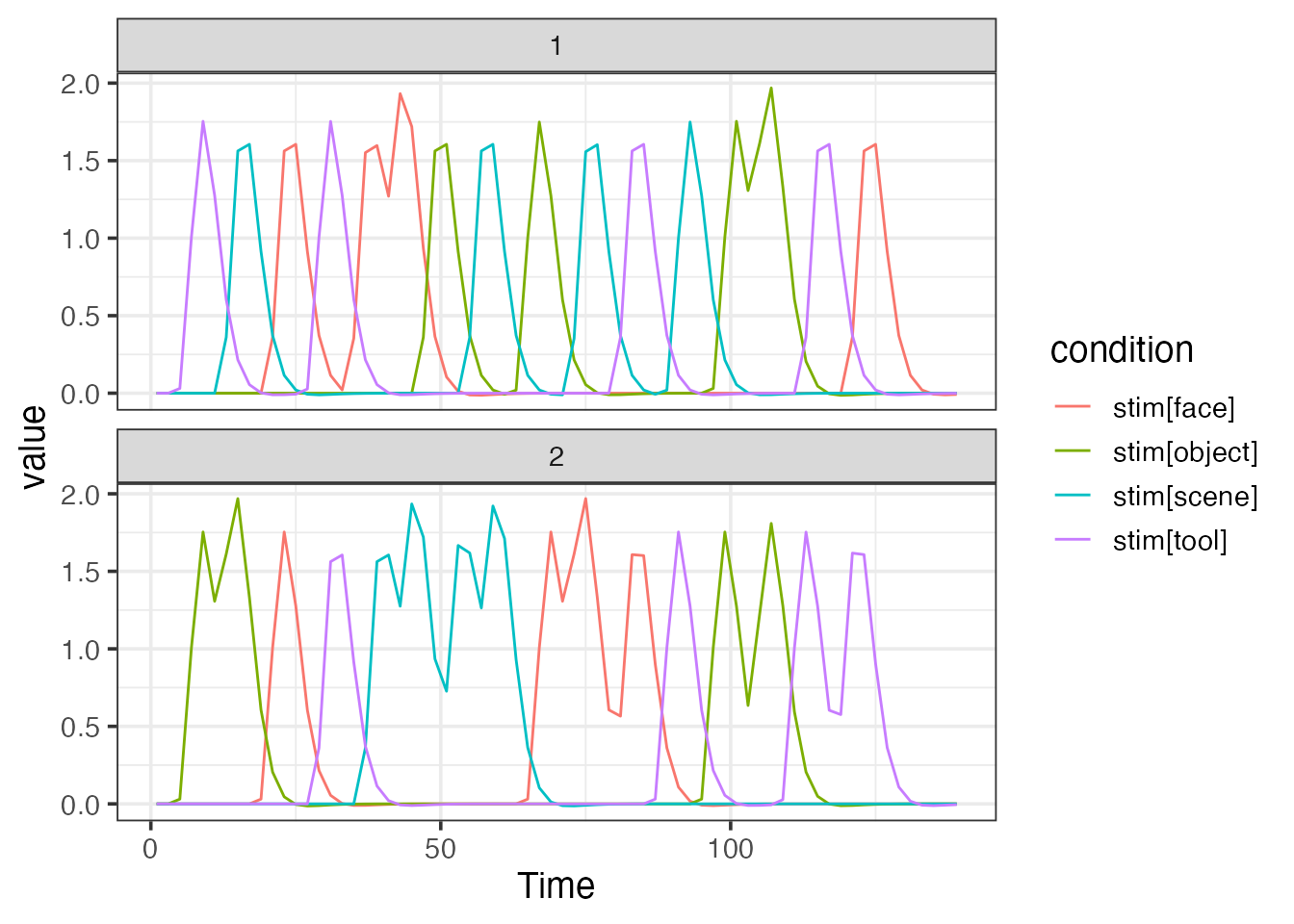
A two-factor fMRI design with multiple blocks
cond1 <- c("face", "scene", "tool", "object")
cond2 <- c("attend", "ignore")
comb <- expand.grid(stim=cond1, task=cond2)
NSTIM <- nrow(comb) * 4
design <- do.call(rbind, lapply(1:2, function(i) {
ind <- sample(rep(1:nrow(comb), length.out=NSTIM))
d <- data.frame(stim=factor(comb$stim[ind]),
task=factor(comb$task[ind]),
ISI=sample(2:6, NSTIM, replace=TRUE),
run=rep(i, NSTIM))
d$onset <- cumsum(d$ISI+2) -2
d
}))
head(design)## stim task ISI run onset
## 1 face ignore 6 1 6
## 2 tool ignore 3 1 11
## 3 tool attend 5 1 18
## 4 object attend 4 1 24
## 5 face attend 5 1 31
## 6 object ignore 3 1 36
## create a 'sampling_frame' describing the temporal outline of the scan consisting of 70 images collected every once every 2 seconds.
TR <- 2
sframe <- sampling_frame(blocklens=c(100,100), TR)
## create the event model d
emodel <- event_model(onset ~ hrf(stim, task), data=design, block = ~ run, sampling_frame=sframe)
print(emodel)##
## ╔══════════════════════════════════════════╗
## ║ fMRI Event Model ║
## ╚══════════════════════════════════════════╝
##
## Model Formula:
## onset ~ hrf(stim, task)
##
## 📈 Model Summary:
## • Number of Terms: 1
## • Total Events: 64
## • Design Matrix Columns: 8
## • Number of Blocks: 2
##
## 📋 Event Table Preview:
## • Variables: stim, task, ISI, run, onset
## stim task ISI run onset
## 1 face ignore 6 1 6
## 2 tool ignore 3 1 11
## 3 tool attend 5 1 18
## ... ( 61 more rows )
##
## 🔍 Model Terms:
##
## Term 1 : stim..task
## • Conditions: stim[face]:task[attend], stim[object]:task[attend], stim[scene]:task[attend], stim[tool]:task[attend], stim[face]:task[ignore], stim[object]:task[ignore], stim[scene]:task[ignore], stim[tool]:task[ignore]
## • HRF Type: spmg1
## • Basis Functions: 1
plot(emodel)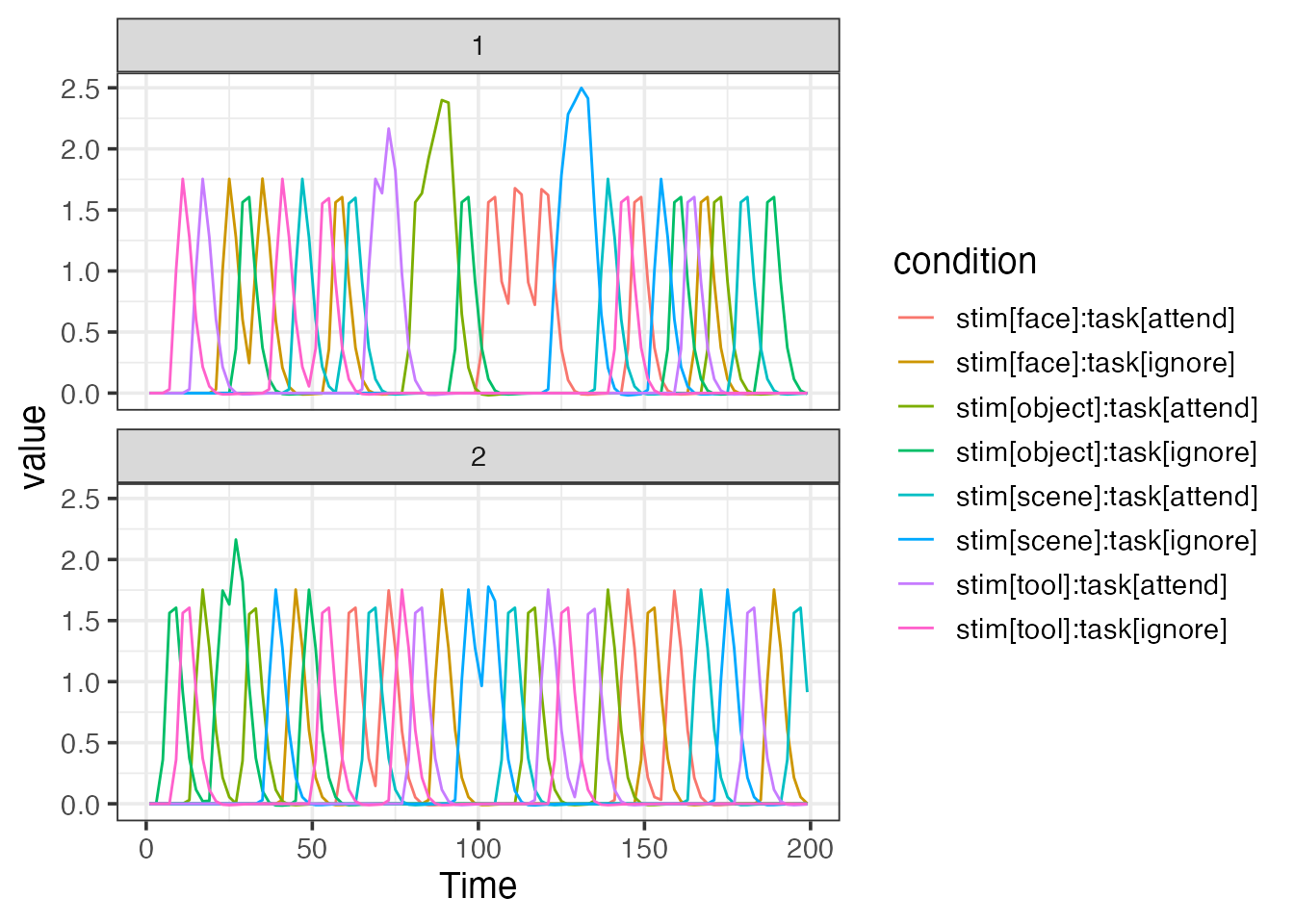
An event-related design with an amplitude-modulated variable
TR <- 2
sframe <- sampling_frame(blocklens=70, TR)
simple_design$RT <- scale(rnorm(nrow(simple_design), mean=700, sd=100))[,1]
## create the event model d
emodel <- event_model(onset ~ hrf(stim) + hrf(RT), data=simple_design, block = ~ run, sampling_frame=sframe)
print(emodel)##
## ╔══════════════════════════════════════════╗
## ║ fMRI Event Model ║
## ╚══════════════════════════════════════════╝
##
## Model Formula:
## onset ~ hrf(stim) + hrf(RT)
##
## 📈 Model Summary:
## • Number of Terms: 2
## • Total Events: 16
## • Design Matrix Columns: 5
## • Number of Blocks: 1
##
## 📋 Event Table Preview:
## • Variables: stim, ISI, run, trial, onset, RT
## stim ISI run trial onset RT
## 1 face 6 1 1 6 -0.4982170
## 2 tool 4 1 2 12 0.6717969
## 3 object 4 1 3 18 -0.1475667
## ... ( 13 more rows )
##
## 🔍 Model Terms:
##
## Term 1 : stim
## • Conditions: stim[face], stim[object], stim[scene], stim[tool]
## • HRF Type: spmg1
## • Basis Functions: 1
##
## Term 2 : RT
## • Conditions: RT
## • HRF Type: spmg1
## • Basis Functions: 1
plot(emodel, "RT")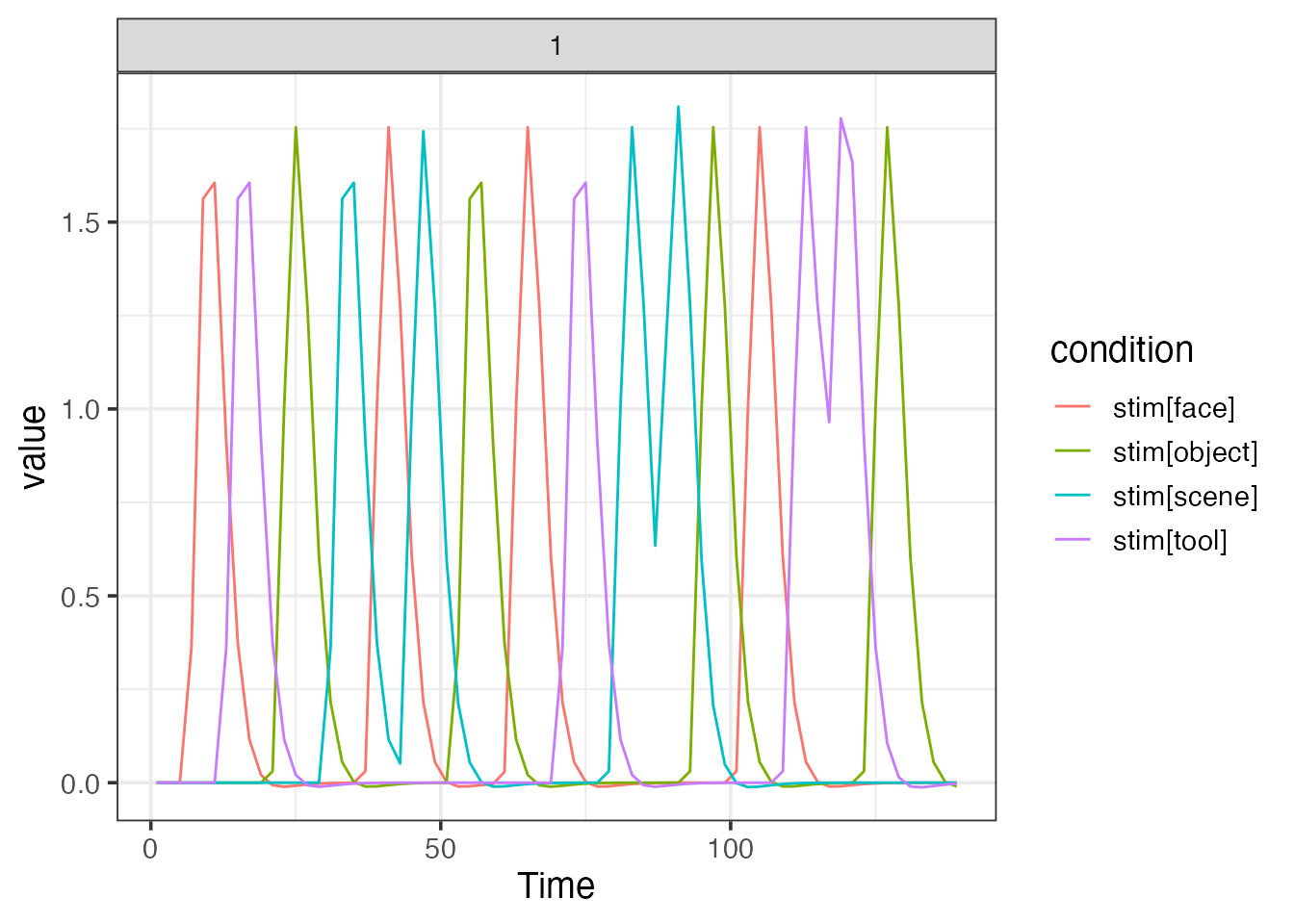
An event-related design with an amplitude-modulated variable crossed with factor
TR <- 2
sframe <- sampling_frame(blocklens=70, TR)
simple_design$RT <- scale(rnorm(nrow(simple_design), mean=700, sd=100))
## create the event model d
emodel <- event_model(onset ~ hrf(stim) + hrf(stim, RT), data=simple_design, block = ~ run, sampling_frame=sframe)
print(emodel)##
## ╔══════════════════════════════════════════╗
## ║ fMRI Event Model ║
## ╚══════════════════════════════════════════╝
##
## Model Formula:
## onset ~ hrf(stim) + hrf(stim, RT)
##
## 📈 Model Summary:
## • Number of Terms: 2
## • Total Events: 16
## • Design Matrix Columns: 8
## • Number of Blocks: 1
##
## 📋 Event Table Preview:
## • Variables: stim, ISI, run, trial, onset, RT
## stim ISI run trial onset RT
## 1 face 6 1 1 6 -0.4525721
## 2 tool 4 1 2 12 1.2814768
## 3 object 4 1 3 18 0.8778785
## ... ( 13 more rows )
##
## 🔍 Model Terms:
##
## Term 1 : stim
## • Conditions: stim[face], stim[object], stim[scene], stim[tool]
## • HRF Type: spmg1
## • Basis Functions: 1
##
## Term 2 : stim..RT
## • Conditions: stim[face]:RT, stim[object]:RT, stim[scene]:RT, stim[tool]:RT
## • HRF Type: spmg1
## • Basis Functions: 1
plot(emodel, "stim:RT")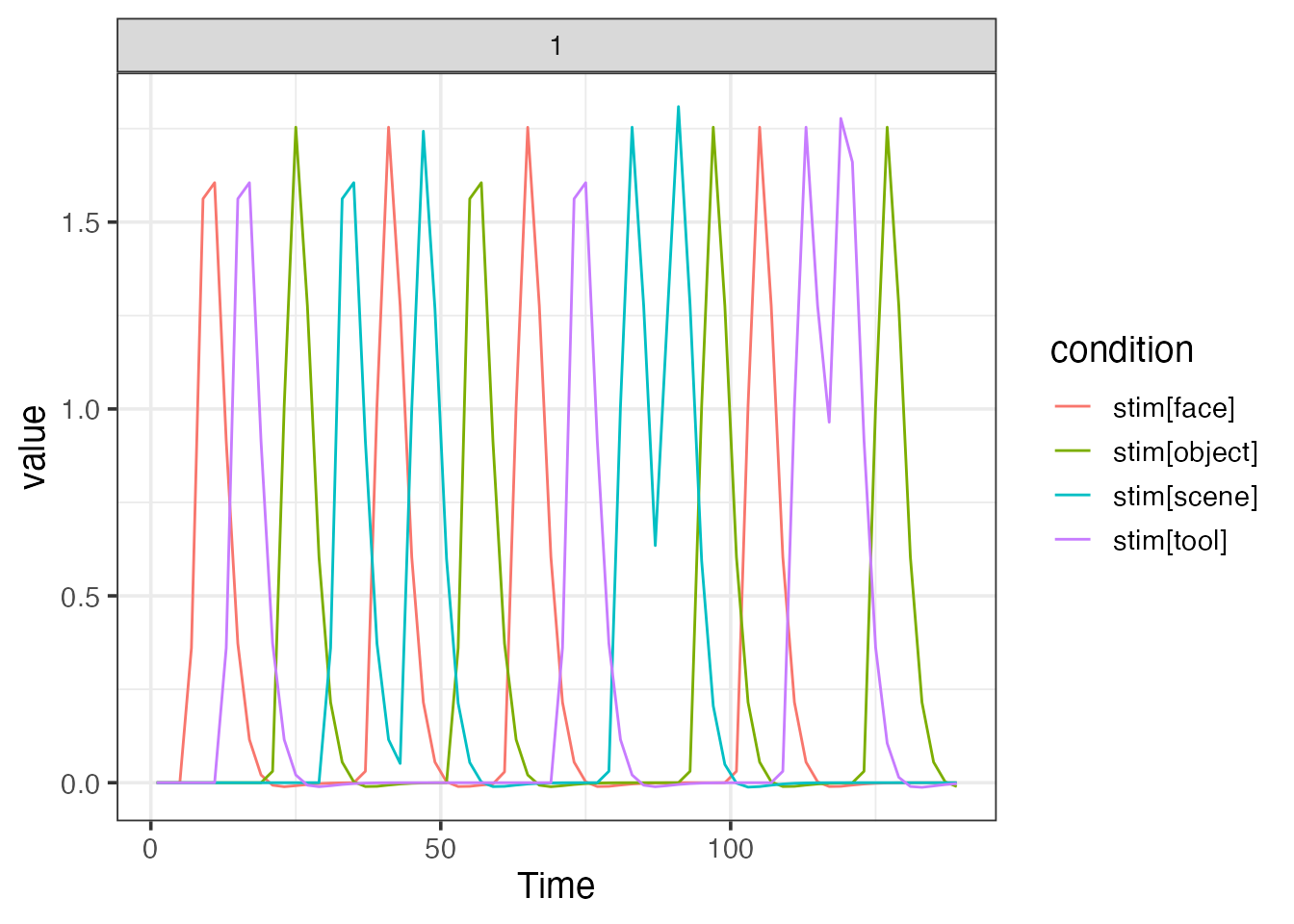
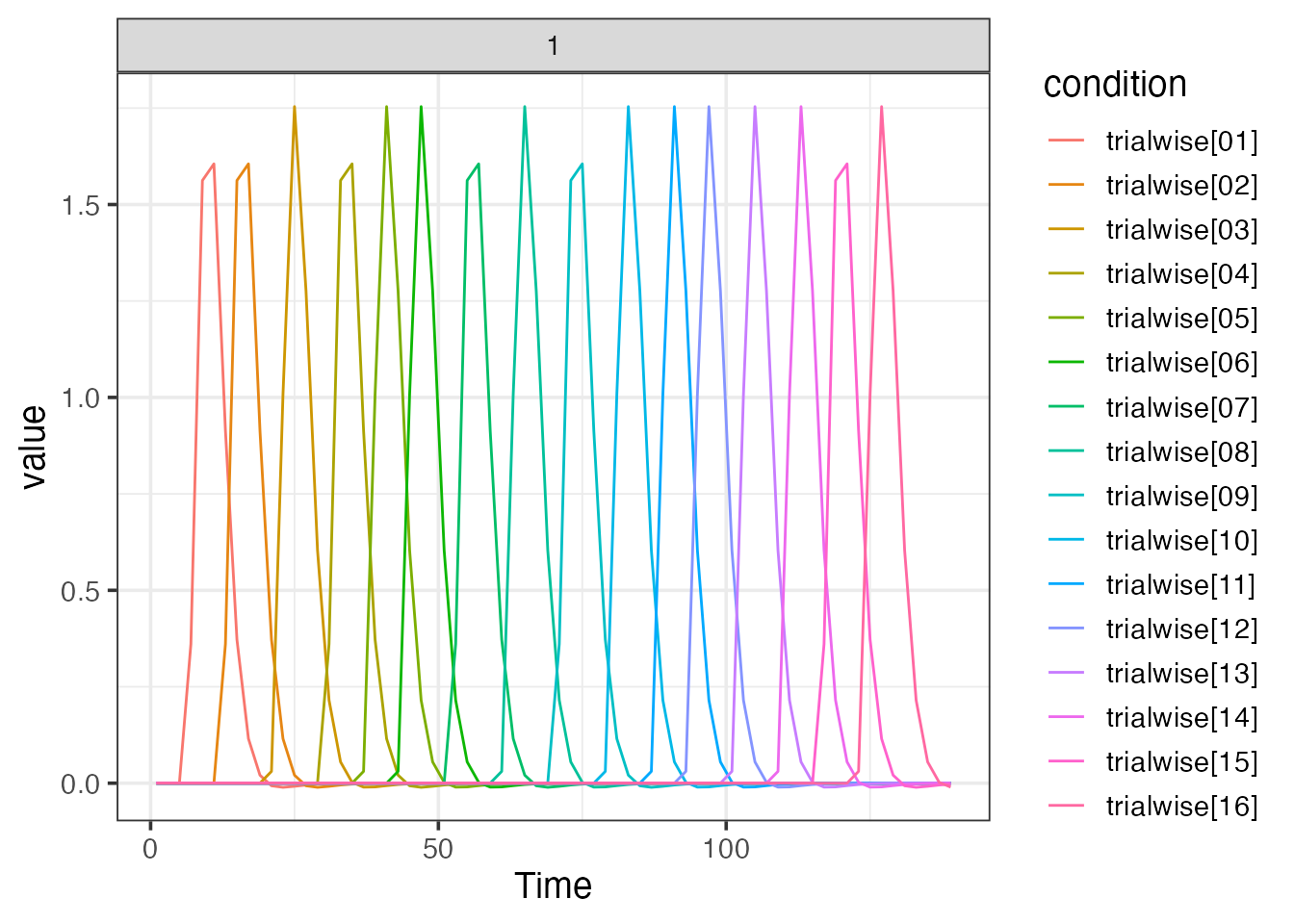
Trialwise event model to construct “beta-series” estimates
TR <- 2
sframe <- sampling_frame(blocklens=70, TR)
# create the event model d
emodel <- event_model(onset ~ trialwise(), data=simple_design, block = ~ run, sampling_frame=sframe)
print(emodel)##
## ╔══════════════════════════════════════════╗
## ║ fMRI Event Model ║
## ╚══════════════════════════════════════════╝
##
## Model Formula:
## onset ~ trialwise()
##
## 📈 Model Summary:
## • Number of Terms: 1
## • Total Events: 16
## • Design Matrix Columns: 16
## • Number of Blocks: 1
##
## 📋 Event Table Preview:
## • Variables: stim, ISI, run, trial, onset, RT
## stim ISI run trial onset RT
## 1 face 6 1 1 6 -0.4525721
## 2 tool 4 1 2 12 1.2814768
## 3 object 4 1 3 18 0.8778785
## ... ( 13 more rows )
##
## 🔍 Model Terms:
##
## Term 1 : trialwise
## • Conditions: trialwise[01], trialwise[02], trialwise[03], trialwise[04], trialwise[05], trialwise[06], trialwise[07], trialwise[08], trialwise[09], trialwise[10], trialwise[11], trialwise[12], trialwise[13], trialwise[14], trialwise[15], trialwise[16]
## • HRF Type: spmg1
## • Basis Functions: 1
plot(emodel, "trialwise")