Baseline Model
Bradley R. Buchsbaum
2023-05-09
Source:vignettes/a_03_baseline_model.Rmd
a_03_baseline_model.RmdfMRI Baseline Model
An “baseline model” refers to the set of regressors used to capture variance associated with noise signals in the fMRI time-series data. These regressors are not formed by convolving a hemodynamic response with event onsets, but rather directly model sources of variance in the data. Low-frequency baseline trends are often modeled with a set of basis functions, such as b-splines, polynomial regressors, or a Fourier basis. Nuisance trends can also be derived from the data itself, for example, the global signal mean, motion parameters, or estimated noise components via PCA or ICA.
Basic baseline model with b-splines and polynomials
TR <- 2
sframe <- sampling_frame(blocklens=c(100,100), TR)
bmodel <- baseline_model(basis="bs", degree=5, sframe=sframe)
bmodel## $drift_term
## $varname
## [1] "baseline_bs_5"
##
## $design_matrix
## # A tibble: 200 × 10
## base_bs1_block_1 base_bs2_block_1 base_bs3_block_1 base_bs4_block_1
## <dbl> <dbl> <dbl> <dbl>
## 1 0 0 0 0
## 2 0.0485 0.000990 0.0000101 0.0000000515
## 3 0.0931 0.00384 0.0000792 0.000000816
## 4 0.134 0.00837 0.000262 0.00000409
## 5 0.171 0.0144 0.000607 0.0000128
## 6 0.205 0.0218 0.00116 0.0000309
## 7 0.236 0.0304 0.00196 0.0000634
## 8 0.264 0.0401 0.00305 0.000116
## 9 0.288 0.0507 0.00446 0.000196
## 10 0.310 0.0621 0.00621 0.000310
## # ℹ 190 more rows
## # ℹ 6 more variables: base_bs5_block_1 <dbl>, base_bs1_block_2 <dbl>,
## # base_bs2_block_2 <dbl>, base_bs3_block_2 <dbl>, base_bs4_block_2 <dbl>,
## # base_bs5_block_2 <dbl>
##
## $colind
## $colind[[1]]
## [1] 1 2 3 4 5
##
## $colind[[2]]
## [1] 6 7 8 9 10
##
##
## $rowind
## $rowind[[1]]
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
## [91] 91 92 93 94 95 96 97 98 99 100
##
## $rowind[[2]]
## [1] 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118
## [19] 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136
## [37] 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154
## [55] 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172
## [73] 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190
## [91] 191 192 193 194 195 196 197 198 199 200
##
##
## attr(,"class")
## [1] "baseline_term" "matrix_term" "fmri_term" "list"
##
## $drift_spec
## $degree
## [1] 5
##
## $basis
## [1] "bs"
##
## $fun
## function (x, df = NULL, knots = NULL, degree = 3, intercept = FALSE,
## Boundary.knots = range(x))
## {
## ord <- 1L + (degree <- as.integer(degree))
## if (ord <= 1)
## stop("'degree' must be integer >= 1")
## nx <- names(x)
## x <- as.vector(x)
## nax <- is.na(x)
## if (nas <- any(nax))
## x <- x[!nax]
## outside <- if (!missing(Boundary.knots)) {
## Boundary.knots <- sort(Boundary.knots)
## (ol <- x < Boundary.knots[1L]) | (or <- x > Boundary.knots[2L])
## }
## else FALSE
## if (!is.null(df) && is.null(knots)) {
## nIknots <- df - ord + (1L - intercept)
## if (nIknots < 0L) {
## nIknots <- 0L
## warning(gettextf("'df' was too small; have used %d",
## ord - (1L - intercept)), domain = NA)
## }
## knots <- if (nIknots > 0L) {
## knots <- seq.int(from = 0, to = 1, length.out = nIknots +
## 2L)[-c(1L, nIknots + 2L)]
## quantile(x[!outside], knots)
## }
## }
## Aknots <- sort(c(rep(Boundary.knots, ord), knots))
## if (any(outside)) {
## warning("some 'x' values beyond boundary knots may cause ill-conditioned bases")
## derivs <- 0:degree
## scalef <- gamma(1L:ord)
## basis <- array(0, c(length(x), length(Aknots) - degree -
## 1L))
## e <- 1/4
## if (any(ol)) {
## k.pivot <- (1 - e) * Boundary.knots[1L] + e * Aknots[ord +
## 1]
## xl <- cbind(1, outer(x[ol] - k.pivot, 1L:degree,
## `^`))
## tt <- splineDesign(Aknots, rep(k.pivot, ord), ord,
## derivs)
## basis[ol, ] <- xl %*% (tt/scalef)
## }
## if (any(or)) {
## k.pivot <- (1 - e) * Boundary.knots[2L] + e * Aknots[length(Aknots) -
## ord]
## xr <- cbind(1, outer(x[or] - k.pivot, 1L:degree,
## `^`))
## tt <- splineDesign(Aknots, rep(k.pivot, ord), ord,
## derivs)
## basis[or, ] <- xr %*% (tt/scalef)
## }
## if (any(inside <- !outside))
## basis[inside, ] <- splineDesign(Aknots, x[inside],
## ord)
## }
## else basis <- splineDesign(Aknots, x, ord)
## if (!intercept)
## basis <- basis[, -1L, drop = FALSE]
## n.col <- ncol(basis)
## if (nas) {
## nmat <- matrix(NA, length(nax), n.col)
## nmat[!nax, ] <- basis
## basis <- nmat
## }
## dimnames(basis) <- list(nx, 1L:n.col)
## a <- list(degree = degree, knots = if (is.null(knots)) numeric(0L) else knots,
## Boundary.knots = Boundary.knots, intercept = intercept)
## attributes(basis) <- c(attributes(basis), a)
## class(basis) <- c("bs", "basis", "matrix")
## basis
## }
## <bytecode: 0x7f993eca6f48>
## <environment: namespace:splines>
##
## $intercept
## [1] "runwise"
##
## $name
## [1] "baseline_bs_5"
##
## attr(,"class")
## [1] "baselinespec" "nuisancespec"
##
## $block_term
## $varname
## [1] "constant"
##
## $blockids
## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2
## [112] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [149] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [186] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
##
## $expanded_blockids
## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2
## [112] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [149] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [186] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## Levels: 1 < 2
##
## $design_matrix
## # A tibble: 200 × 2
## constant_1 constant_2
## <dbl> <dbl>
## 1 1 0
## 2 1 0
## 3 1 0
## 4 1 0
## 5 1 0
## 6 1 0
## 7 1 0
## 8 1 0
## 9 1 0
## 10 1 0
## # ℹ 190 more rows
##
## $nblocks
## [1] 2
##
## $colind
## $colind[[1]]
## [1] 1
##
## $colind[[2]]
## [1] 2
##
##
## $rowind
## $rowind$`1`
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
## [91] 91 92 93 94 95 96 97 98 99 100
##
## $rowind$`2`
## [1] 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118
## [19] 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136
## [37] 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154
## [55] 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172
## [73] 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190
## [91] 191 192 193 194 195 196 197 198 199 200
##
##
## attr(,"class")
## [1] "block_term" "matrix_term" "fmri_term" "list"
##
## $nuisance_term
## NULL
##
## $sampling_frame
## $blocklens
## [1] 100 100
##
## $TR
## [1] 2 2
##
## $start_time
## [1] 1 1
##
## $blockids
## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2
## [112] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [149] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [186] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
##
## $time
## [1] 1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35
## [19] 37 39 41 43 45 47 49 51 53 55 57 59 61 63 65 67 69 71
## [37] 73 75 77 79 81 83 85 87 89 91 93 95 97 99 101 103 105 107
## [55] 109 111 113 115 117 119 121 123 125 127 129 131 133 135 137 139 141 143
## [73] 145 147 149 151 153 155 157 159 161 163 165 167 169 171 173 175 177 179
## [91] 181 183 185 187 189 191 193 195 197 199 1 3 5 7 9 11 13 15
## [109] 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 47 49 51
## [127] 53 55 57 59 61 63 65 67 69 71 73 75 77 79 81 83 85 87
## [145] 89 91 93 95 97 99 101 103 105 107 109 111 113 115 117 119 121 123
## [163] 125 127 129 131 133 135 137 139 141 143 145 147 149 151 153 155 157 159
## [181] 161 163 165 167 169 171 173 175 177 179 181 183 185 187 189 191 193 195
## [199] 197 199
##
## $precision
## [1] 0.1
##
## attr(,"class")
## [1] "sampling_frame" "list"
##
## attr(,"class")
## [1] "baseline_model" "list"
plot(bmodel)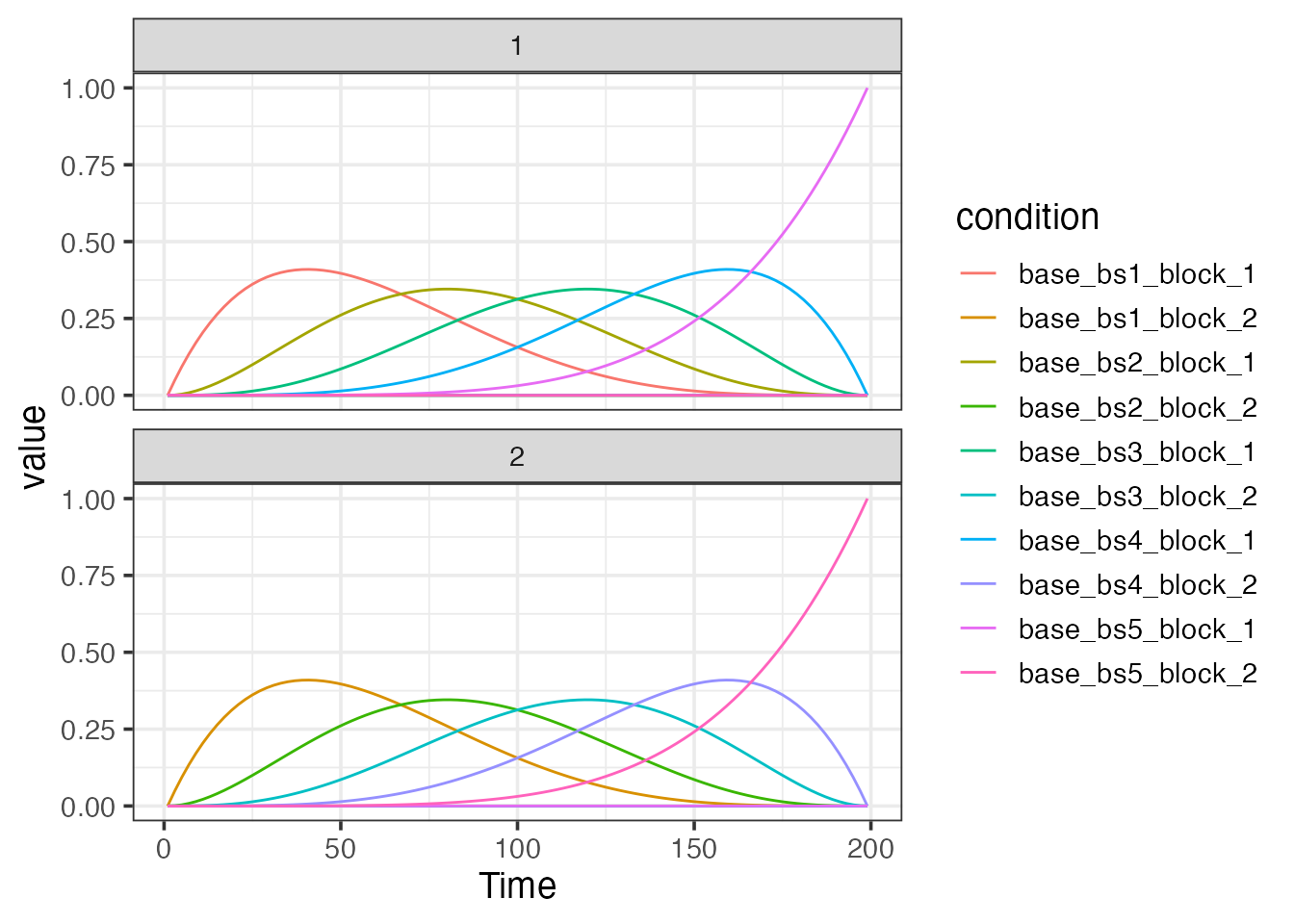
bmodel2 <- baseline_model(basis="poly", degree=5, sframe=sframe)
plot(bmodel2)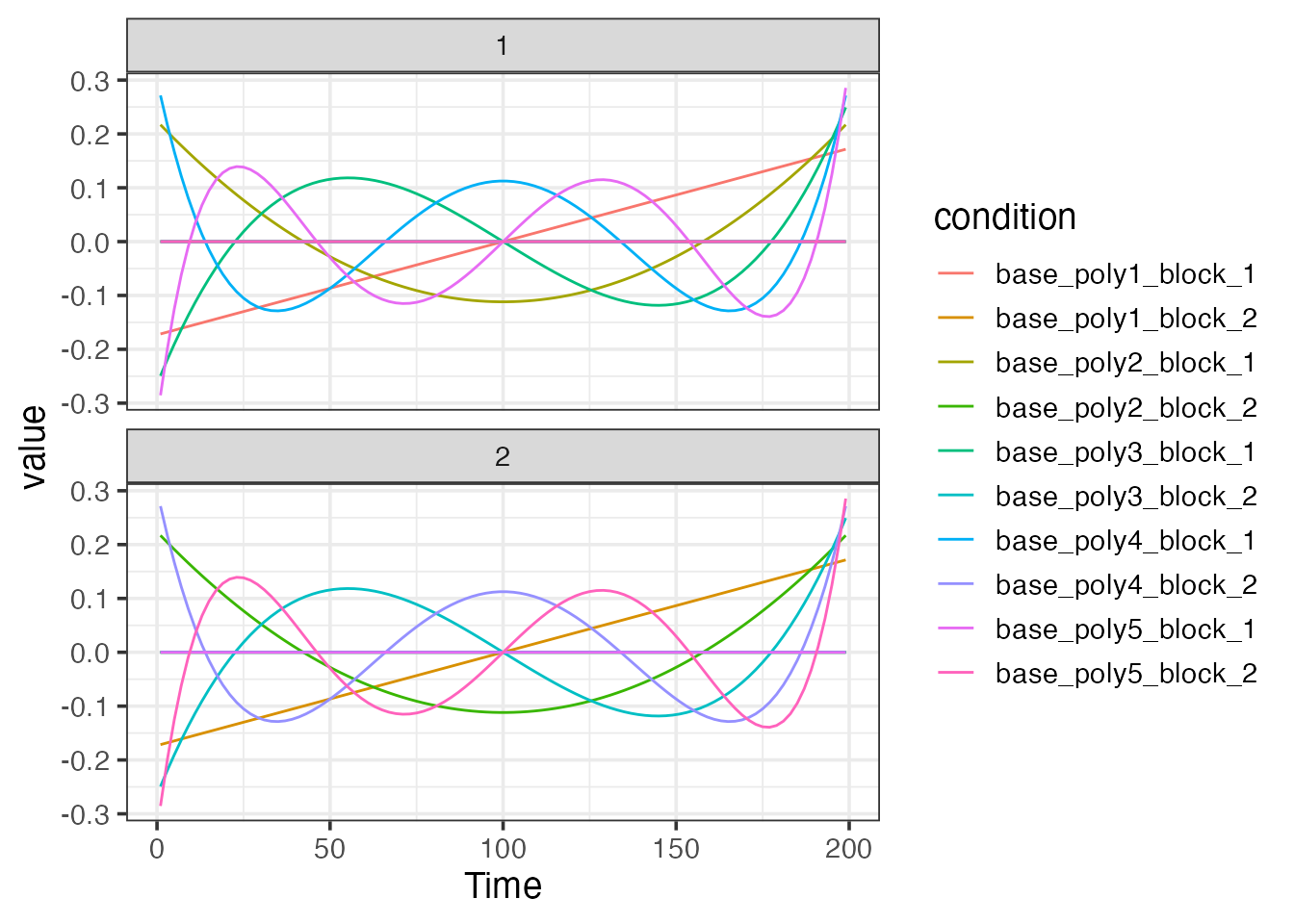
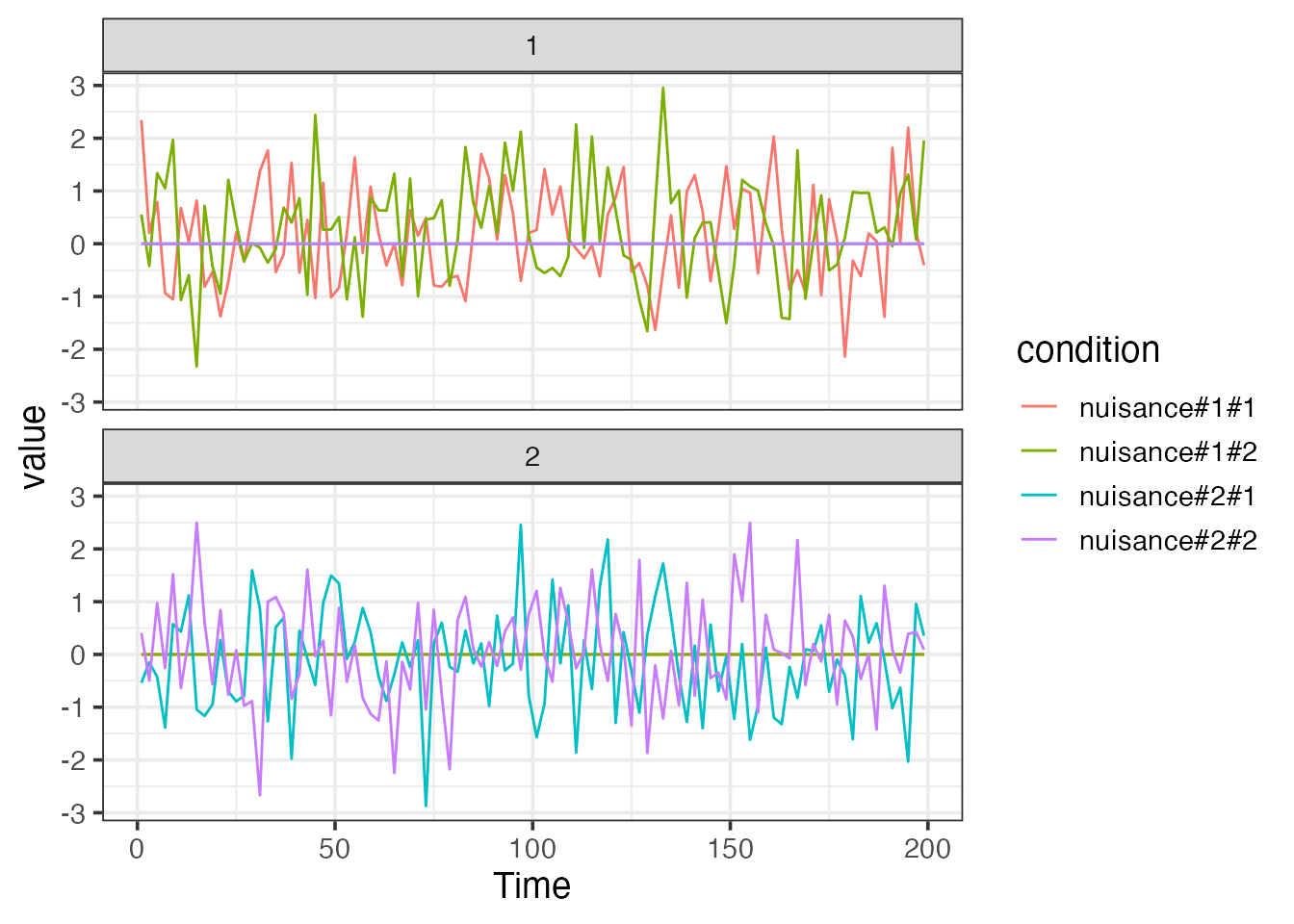
Adding arbitrary nuisance regressors
In addition to structured basis sets, we can also add arbitrary
signals to the baseline model. Such signals are also organized by block
on the assumption that nuisance signals are often block-specific and
“reset” across scans. To add a set of nusiance variables to the baseline
model, then, they should be split by block and placed in a
list.
TR <- 2
sframe <- sampling_frame(blocklens=c(100,100), TR)
## generate two arbitrary nusiance signals.
nuis1 <- rnorm(100*2)
nuis2 <- rnorm(100*2)
## construct a set of data.frames, each with two columns (nuis1 and nuis2)
nuislist <- split(as.data.frame(cbind(nuis1, nuis2)), blockids(sframe))
bmodel <- baseline_model(basis="bs", degree=5, sframe=sframe, nuisance_list=nuislist)
print(terms(bmodel))## $constant
## $varname
## [1] "constant"
##
## $blockids
## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2
## [112] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [149] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [186] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
##
## $expanded_blockids
## [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
## [75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2
## [112] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [149] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [186] 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## Levels: 1 < 2
##
## $design_matrix
## # A tibble: 200 × 2
## constant_1 constant_2
## <dbl> <dbl>
## 1 1 0
## 2 1 0
## 3 1 0
## 4 1 0
## 5 1 0
## 6 1 0
## 7 1 0
## 8 1 0
## 9 1 0
## 10 1 0
## # ℹ 190 more rows
##
## $nblocks
## [1] 2
##
## $colind
## $colind[[1]]
## [1] 1
##
## $colind[[2]]
## [1] 2
##
##
## $rowind
## $rowind$`1`
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
## [91] 91 92 93 94 95 96 97 98 99 100
##
## $rowind$`2`
## [1] 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118
## [19] 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136
## [37] 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154
## [55] 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172
## [73] 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190
## [91] 191 192 193 194 195 196 197 198 199 200
##
##
## attr(,"class")
## [1] "block_term" "matrix_term" "fmri_term" "list"
##
## $baseline_bs_5
## $varname
## [1] "baseline_bs_5"
##
## $design_matrix
## # A tibble: 200 × 10
## base_bs1_block_1 base_bs2_block_1 base_bs3_block_1 base_bs4_block_1
## <dbl> <dbl> <dbl> <dbl>
## 1 0 0 0 0
## 2 0.0485 0.000990 0.0000101 0.0000000515
## 3 0.0931 0.00384 0.0000792 0.000000816
## 4 0.134 0.00837 0.000262 0.00000409
## 5 0.171 0.0144 0.000607 0.0000128
## 6 0.205 0.0218 0.00116 0.0000309
## 7 0.236 0.0304 0.00196 0.0000634
## 8 0.264 0.0401 0.00305 0.000116
## 9 0.288 0.0507 0.00446 0.000196
## 10 0.310 0.0621 0.00621 0.000310
## # ℹ 190 more rows
## # ℹ 6 more variables: base_bs5_block_1 <dbl>, base_bs1_block_2 <dbl>,
## # base_bs2_block_2 <dbl>, base_bs3_block_2 <dbl>, base_bs4_block_2 <dbl>,
## # base_bs5_block_2 <dbl>
##
## $colind
## $colind[[1]]
## [1] 1 2 3 4 5
##
## $colind[[2]]
## [1] 6 7 8 9 10
##
##
## $rowind
## $rowind[[1]]
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
## [91] 91 92 93 94 95 96 97 98 99 100
##
## $rowind[[2]]
## [1] 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118
## [19] 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136
## [37] 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154
## [55] 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172
## [73] 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190
## [91] 191 192 193 194 195 196 197 198 199 200
##
##
## attr(,"class")
## [1] "baseline_term" "matrix_term" "fmri_term" "list"
##
## $nuisance
## $varname
## [1] "nuisance"
##
## $design_matrix
## # A tibble: 200 × 4
## `nuisance#1#1` `nuisance#1#2` `nuisance#2#1` `nuisance#2#2`
## <dbl> <dbl> <dbl> <dbl>
## 1 -0.326 0.0602 0 0
## 2 1.25 -1.59 0 0
## 3 -0.135 1.37 0 0
## 4 -1.42 -0.111 0 0
## 5 0.310 -0.410 0 0
## 6 -1.01 -1.12 0 0
## 7 0.561 -0.0445 0 0
## 8 -1.88 2.24 0 0
## 9 0.747 -0.760 0 0
## 10 0.439 0.396 0 0
## # ℹ 190 more rows
##
## $colind
## $colind[[1]]
## [1] 1 2
##
## $colind[[2]]
## [1] 3 4
##
##
## $rowind
## $rowind$`1`
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
## [91] 91 92 93 94 95 96 97 98 99 100
##
## $rowind$`2`
## [1] 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118
## [19] 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136
## [37] 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154
## [55] 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172
## [73] 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190
## [91] 191 192 193 194 195 196 197 198 199 200
##
##
## attr(,"class")
## [1] "baseline_term" "matrix_term" "fmri_term" "list"
plot(bmodel, term_name="nuisance")