OASIS Theory: Algebra and Implementation Details
fmrilss Development Team
2025-10-31
oasis_theory.RmdMotivation
Optimized Analytic Single-pass Inverse Solution (OASIS) extends Least Squares Separate (LSS) estimation through algebraic reformulation that enables single-pass computation of all trial estimates. For a single-basis HRF (K = 1) without ridge, OASIS reduces exactly to the closed-form LSS solution; the per‑trial 2x2 normal equations are the same. The value of OASIS is in batching those solves efficiently and generalizing the same algebra to multi‑basis HRFs (2Kx2K) with optional ridge and diagnostics. This document provides the mathematical foundation and implementation details.
Computational Intuition
Standard LSS requires N separate GLM fits for N trials, each involving: 1. Matrix assembly: O(T²) operations 2. QR decomposition: O(T³) operations 3. Back-substitution: O(T²) operations
Total complexity: O(NT³) for N trials
OASIS recognizes that these N models share substantial structure. By factoring out common computations, OASIS reduces complexity to: 1. Single QR decomposition: O(T³) 2. Shared projections: O(NT²) 3. Per-trial solutions: O(N)
Total complexity: O(T³ + NT²), a significant reduction when N is large.
Visual Comparison of Computational Scaling
# Demonstrate computational scaling
N_trials <- c(10, 50, 100, 200, 500, 1000)
T_points <- 200 # Fixed number of timepoints
# Simplified complexity models (arbitrary units)
classical_ops <- N_trials * T_points^3 / 1e6 # O(NT³)
oasis_ops <- (T_points^3 + N_trials * T_points^2) / 1e6 # O(T³ + NT²)
# Create comparison plot
plot(N_trials, classical_ops, type='l', col='red', lwd=2,
xlab='Number of Trials', ylab='Computational Operations (millions)',
main='Computational Complexity: Classical LSS vs OASIS',
ylim=c(0, max(classical_ops)))
lines(N_trials, oasis_ops, col='blue', lwd=2)
legend('topleft', c('Classical LSS', 'OASIS'),
col=c('red', 'blue'), lwd=2, bty='n')
# Add shaded region showing computational savings
polygon(c(N_trials, rev(N_trials)),
c(classical_ops, rev(oasis_ops)),
col=rgb(0.2, 0.8, 0.2, 0.3), border=NA)
text(500, mean(c(classical_ops[4], oasis_ops[4])),
'Computational\nSavings', col='darkgreen')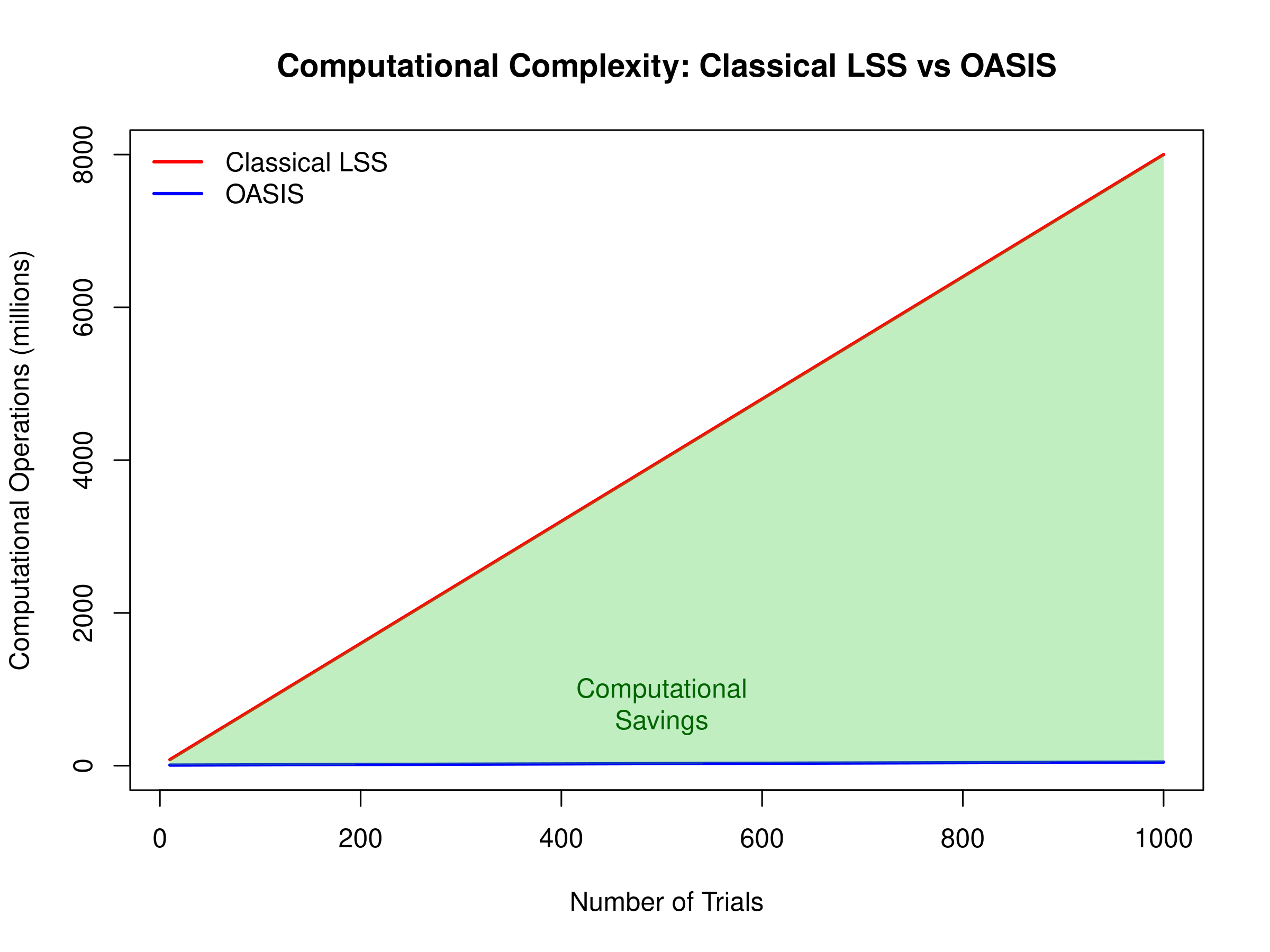
Computational complexity: Classical LSS vs OASIS
Prerequisites
This vignette assumes familiarity with: - QR decomposition and orthogonal projection matrices - Ridge regression and regularization - Matrix calculus and linear algebra - The standard LSS formulation
Code references point to R/oasis_glue.R and
src/oasis_core.cpp implementations.
Notation used throughout:
- : voxel data ( time points, voxels)
- : trial regressors for one condition
- : nuisance/experimental regressors shared across trials
- : orthogonal projector removing nuisance effects ( comes from QR factorisation of )
- Inner products are denoted
We first treat the single-basis case (one regressor per trial) before generalizing to multi-basis HRFs.
Classical LSS Recap
Classical LSS fits, for each trial , a GLM with design , where . Solving each model independently costs QR factorizations. Algebraically, the trial-specific beta can be expressed as
OASIS extracts and reuses the common computational components (projections, norms, cross-products) across all trials, computing each only once.
Single-Basis OASIS Algebra
After residualising against nuisance regressors we define:
Let and . The pair solving the 2x2 system for trial and voxel is obtained from
with ridge penalties
.
The inverse of
is analytic, so
This is exactly what oasis_betas_closed_form()
implements (C++ file src/oasis_core.cpp). The
precomputation step oasis_precompute_design() produces
once, while oasis_AtY_SY_blocked() streams through voxels
to obtain
and
.
Multi-Basis Extension
When the HRF contributes basis functions, each trial has columns . Define
Per voxel we need (stacked blocks of size ) and . The block system is
where
.
oasisk_betas() solves this 2Kx2K system via Cholesky
factorisation. Ridge again adds
and
to the block diagonals. Compared to the single-basis path, only the
shapes of the cached matrices differ; the solve is still analytic per
trial/voxel block.
The companion oasisk_betas_se() extends the SSE/variance
calculation to the multi-basis case, using the same building blocks.
HRF-Aware Design Construction
OASIS can construct
on the fly from event specifications.
.oasis_build_X_from_events() uses
fmrihrf::regressor_set() to generate trial-wise columns
(and optional other-condition aggregates) given:
-
cond$onsets: per-trial onset times -
cond$hrf: HRF object (canonical, FIR, multi-basis, user-defined) -
cond$span,precision,method: convolution controls
This design is then residualised against nuisance regressors and fed
into the algebra above. Because the HRF definition enters directly,
switching HRFs or running grid searches automatically regenerates a
matching design. When you provide an explicit X, OASIS
skips this step and assumes you have already encoded the HRF in the
matrix.
AR(1) Whitening
oasis$whiten = "ar1" estimates a common AR(1)
coefficient from residualised data. .oasis_ar1_whitener()
computes
and applies Toeplitz-safe differencing:
The same transformation is applied to and nuisance regressors before the standard OASIS algebra runs. Whitening preserves the single-pass benefits because the transformed data are treated exactly like the original inputs.
Diagnostics Output
When oasis$return_diag = TRUE, OASIS returns the
precomputed design scalars:
- Single-basis:
(from
oasis_precompute_design()) - Multi-basis:
(from
oasisk_precompute_design())
These matrices are useful for checking trial collinearity, energy, and the effect of ridge scaling.
Algorithm Summary
Putting everything together, the single-basis solver proceeds as follows:
- Residualise and against nuisance regressors, optionally with whitening.
- Compute
(
oasis_precompute_design). - Stream through voxels in blocks, forming
and
(
oasis_AtY_SY_blocked). - Apply ridge scaling (absolute or fractional) to obtain .
- For each trial, evaluate the closed-form (and if SEs requested).
- Optionally compute SEs and diagnostics.
The multi-basis path swaps steps 2–5 for their block equivalents. In both cases, the cost is dominated by the single projection of and the matrix–vector multiplies in step 3, giving complexity with a small trial-dependent overhead.
Complexity and Memory
- Projection / whitening: arithmetic, memory for confounds
- Precomputation:
- Products (blocked): with block size tuning
- Closed-form solves: with negligible constants (2x2 or 2Kx2K systems)
Compared to classical LSS ( separate regressions), OASIS shaves off repeated projections and linear solves, yielding substantial speedups when or is large.
References
- Mumford, J. A., Turner, B. O., Ashby, F. G., & Poldrack, R. A. (2012). Deconvolving BOLD activation in event-related designs for multivoxel pattern classification analyses. NeuroImage, 59(3), 2636–2643.
- fmrilss source files
R/oasis_glue.Randsrc/oasis_core.cpp(for implementation alignment).
sessionInfo()
#> R version 4.5.1 (2025-06-13)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> loaded via a namespace (and not attached):
#> [1] digest_0.6.37 desc_1.4.3 R6_2.6.1 fastmap_1.2.0
#> [5] xfun_0.53 cachem_1.1.0 knitr_1.50 htmltools_0.5.8.1
#> [9] rmarkdown_2.30 lifecycle_1.0.4 cli_3.6.5 pkgdown_2.1.3
#> [13] sass_0.4.10 textshaping_1.0.4 jquerylib_0.1.4 systemfonts_1.3.1
#> [17] compiler_4.5.1 tools_4.5.1 ragg_1.5.0 evaluate_1.0.5
#> [21] bslib_0.9.0 yaml_2.3.10 jsonlite_2.0.0 rlang_1.1.6
#> [25] fs_1.6.6