Edge-Preserving Guided Filtering (3D/4D)
fmrismooth authors
2025-08-28
guided-filter.RmdGuided filtering is a fast, edge‑preserving smoother. Unlike bilateral filtering, it assumes a local linear model between the output and a guide image within each window. The regularization parameter prevents overfitting across noisy regions and avoids edge leakage.
This vignette shows how to use the 3D and 4D guided filters and how to choose their parameters.
Spatial guided filtering (3D)
The 3D variant processes one volume using a sliding window. The
window radius controls spatial scale; the regularization
eps sets how “rigid” the fit is to the guide: small values
track edges more closely, larger values behave more like an averaging
filter.
d3 <- c(10, 10, 10)
vol <- array(100, dim = d3)
vol[6:10, , ] <- vol[6:10, , ] + 20
noisy <- vol + array(rnorm(prod(d3), sd = 6), dim = d3)
gf_smooth <- guided_filter3d(noisy, radius = 2L, eps = 0.5^2)
gf_rigid <- guided_filter3d(noisy, radius = 2L, eps = 1.5^2)
c(var_noisy = var(as.vector(noisy)),
var_smooth = var(as.vector(gf_smooth)),
var_rigid = var(as.vector(gf_rigid)))
#> var_noisy var_smooth var_rigid
#> 137.3983 136.9045 133.1892
zmid <- ceiling(d3[3]/2)
viz3 <- rbind(
slice_df3d(noisy, zmid, "noisy"),
slice_df3d(gf_smooth, zmid, "eps=0.25"),
slice_df3d(gf_rigid, zmid, "eps=2.25")
)
ggplot(viz3, aes(x, y, fill = val)) + geom_raster() + coord_fixed() + scale_y_reverse() +
scale_fill_gradient(low = "black", high = "white") + facet_wrap(~method) + theme_minimal(base_size = 10) +
labs(title = "3D guided filter: mid-slice", x = NULL, y = NULL, fill = "intensity")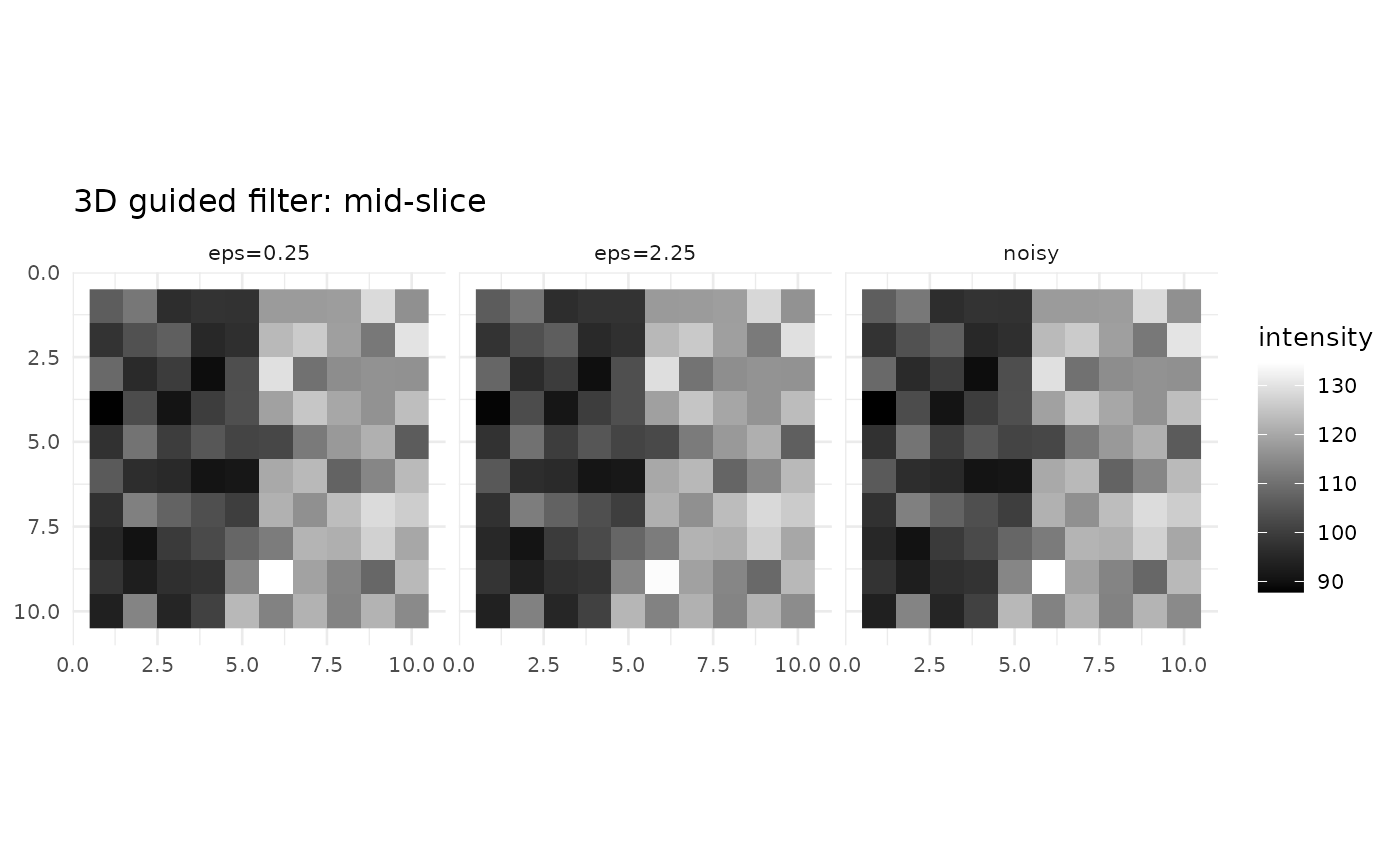
Using the input itself as the guide is common. You can also pass an external guide, for example a T1 anatomical, to steer the filter across low‑contrast fMRI regions.
Spatiotemporal guided filtering (4D)
The 4D variant applies the spatial guided filter to each frame, then
an optional temporal guided step per voxel. The temporal radius sets how
many frames on either side to include; eps_t plays the same
role as in space. Motion‑aware temporal weights can down‑weight
transitions between frames with large motion changes.
d4 <- c(8, 8, 8, 14)
x <- array(100, dim = d4) + array(rnorm(prod(d4), sd = 5), dim = d4)
out_no_temp <- guided_filter4d(x, radius_sp = 2L, eps_sp = 0.6^2, radius_t = 0L)
out_temp <- guided_filter4d(x, radius_sp = 2L, eps_sp = 0.6^2, radius_t = 1L, eps_t = 0.3^2)
c(var_no_temp = var(as.vector(out_no_temp)),
var_temp = var(as.vector(out_temp)))
#> var_no_temp var_temp
#> 25.00629 24.83296
zmid <- ceiling(d4[3]/2); tmid <- ceiling(d4[4]/2)
viz4 <- rbind(
slice_df4d(x, zmid, tmid, "noisy"),
slice_df4d(out_no_temp, zmid, tmid, "radius_t=0"),
slice_df4d(out_temp, zmid, tmid, "radius_t=1")
)
ggplot(viz4, aes(x, y, fill = val)) + geom_raster() + coord_fixed() + scale_y_reverse() +
scale_fill_gradient(low = "black", high = "white") + facet_wrap(~method) + theme_minimal(base_size = 10) +
labs(title = "4D guided filter: mid-slice/frame", x = NULL, y = NULL, fill = "intensity")
When you have per‑frame motion parameters, supply them as
motion_params; the filter will dampen temporal averaging
across high‑motion transitions.