Building fMRI Regressors
Bradley R. Buchsbaum
2026-02-20
Source:vignettes/a_02_regressor.Rmd
a_02_regressor.RmdIntroduction: What is a Regressor?
In fMRI analysis, a regressor (or predictor) represents the expected BOLD signal timecourse associated with a specific experimental condition or event type. It’s typically created by convolving a series of event onsets (often represented as delta functions or “sticks”) with a hemodynamic response function (HRF).
fmrihrf provides the regressor() function
to easily create these objects from event timings and an HRF. While
these regressor objects are often constructed automatically by modeling
functions in other packages, this vignette explores how to create and
manipulate them directly, offering finer control over the model
components.
Basic Regressor from Event Onsets
Suppose we have a simple event-related fMRI design with stimuli
presented every 12 seconds. We want to model these events using the SPM
canonical HRF (HRF_SPMG1). The events are brief, so we
model them with a duration of 0 seconds (instantaneous).
# Define event onsets
onsets <- seq(0, 10 * 12, by = 12)
# Create the regressor object
# Uses HRF_SPMG1 by default if no hrf is specified
# Duration is 0 by default
reg1 <- regressor(onsets = onsets, hrf = HRF_SPMG1)
# Print the regressor object to see its properties (uses new print.Reg method)
print(reg1)
# Access components using helper functions
head(onsets(reg1))
#> [1] 0 12 24 36 48 60
nbasis(reg1)
#> [1] 1Evaluating and Plotting a Regressor
A regressor object stores the event information but
doesn’t automatically compute the timecourse. To get the predicted BOLD
signal at specific time points (e.g., corresponding to scan acquisition
times), we use the evaluate() function.
# Define a time grid corresponding to scan times (e.g., TR=2s)
TR <- 2
scan_times <- seq(0, 140, by = TR)
# Plot the regressor using the plot() method
# This automatically evaluates and plots with onset markers
plot(reg1, grid = scan_times)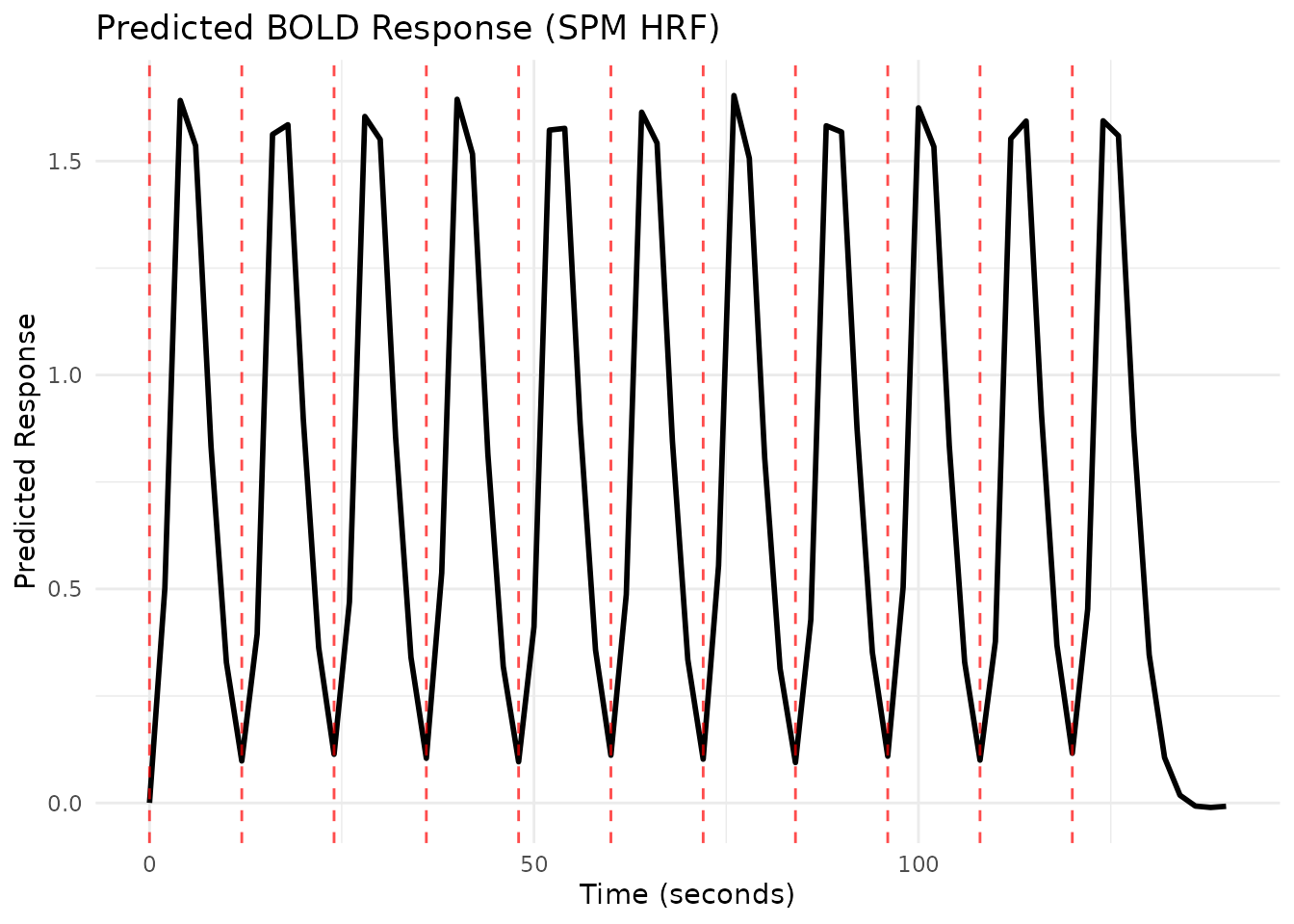
Varying Event Durations
Sometimes events have different durations. The duration
argument in regressor() can take a vector matching the
length of onsets.
# Example onsets and durations
onsets_var_dur <- seq(0, 5 * 12, length.out = 6)
durations_var <- 1:length(onsets_var_dur) # Durations increase from 1s to 6s
# Create regressor with varying durations
reg_var_dur <- regressor(onsets_var_dur, HRF_SPMG1, duration = durations_var)
print(reg_var_dur)
# Plot the regressor
scan_times_dur <- seq(0, max(onsets_var_dur) + 30, by = TR)
plot(reg_var_dur, grid = scan_times_dur)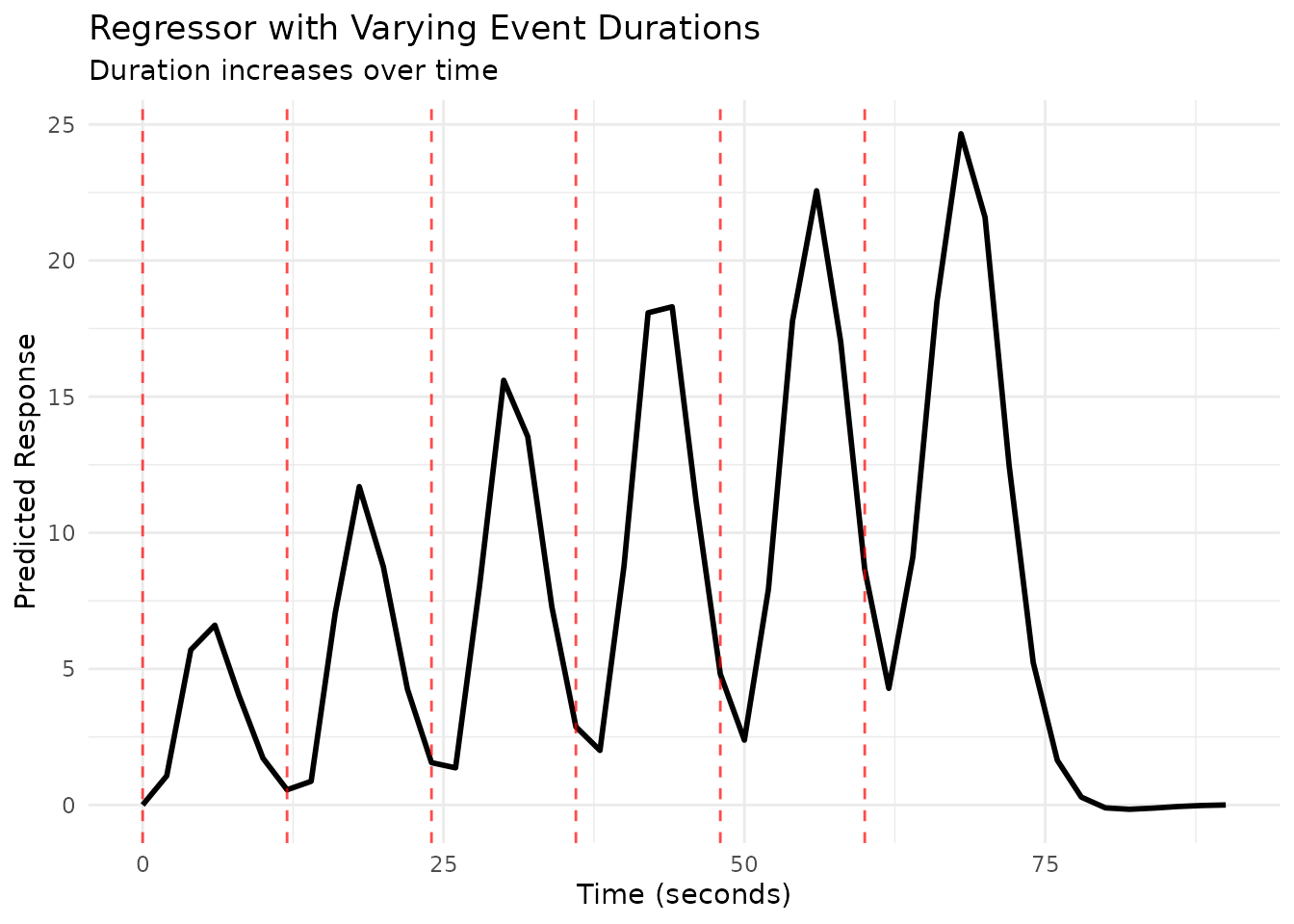
Duration and Summation
By default (summate=TRUE), the predicted response
accumulates if events overlap or have extended duration. Setting
summate=FALSE preserves the same temporal profile but the
peak amplitude does not grow with duration.
# Create regressor with varying durations, summate=FALSE
reg_var_dur_nosum <- regressor(onsets_var_dur, HRF_SPMG1,
duration = durations_var, summate = FALSE)
# Compare summating vs non-summating using plot_regressors()
plot_regressors(reg_var_dur, reg_var_dur_nosum,
labels = c("summate=TRUE", "summate=FALSE"),
grid = scan_times_dur,
title = "Effect of Summation on Response",
subtitle = "Same events with varying durations")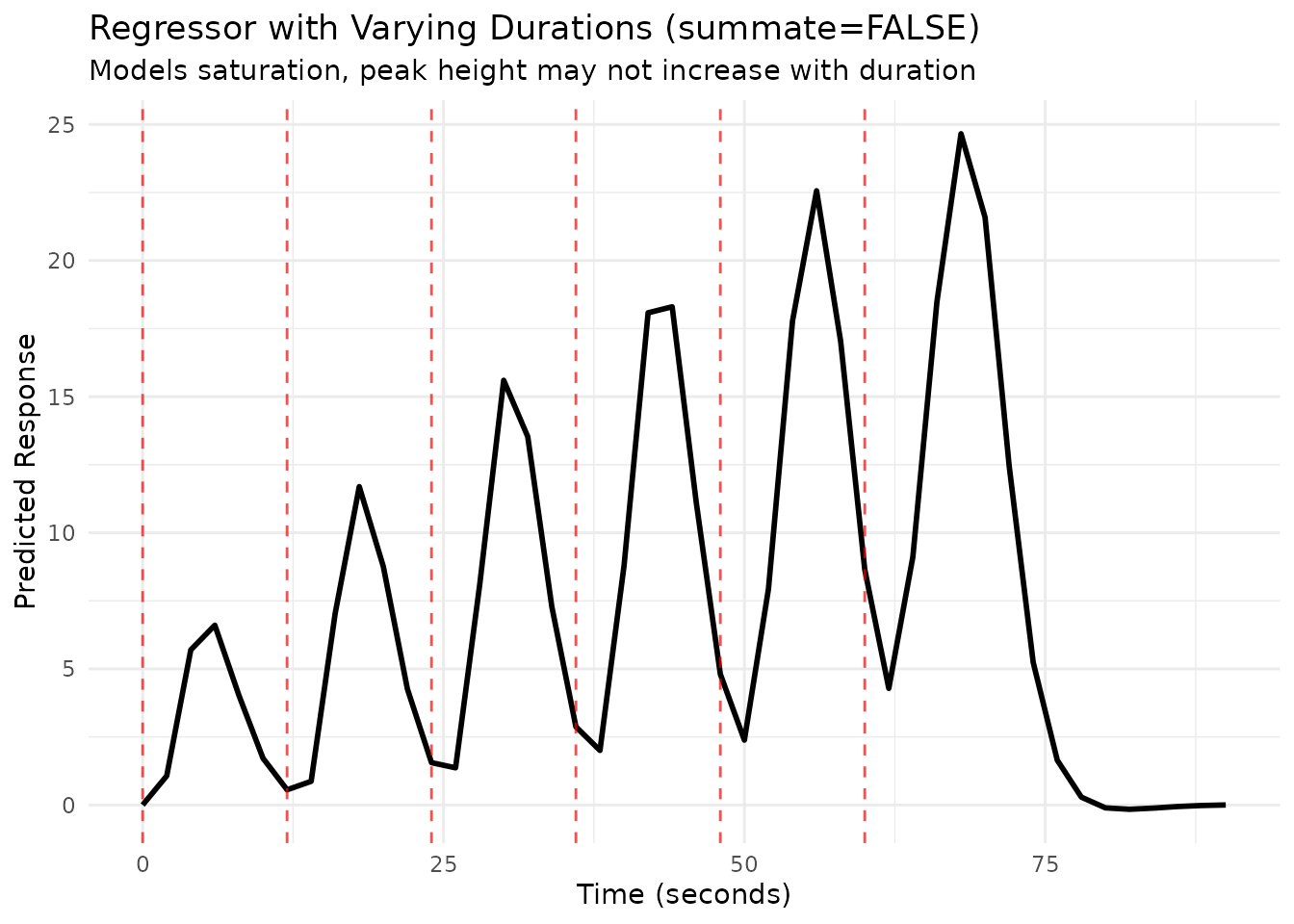
Varying Event Amplitudes (Parametric Modulation)
We can model variations in event intensity or some associated
parameter by providing an amplitude vector. This creates a
parametric regressor where the height of the HRF for each event
is scaled by the corresponding amplitude value.
# Example onsets and amplitudes (e.g., representing task difficulty)
onsets_amp <- seq(0, 10 * 12, length.out = 11)
amplitudes_raw <- 1:length(onsets_amp)
# It's common practice to center the modulator
amplitudes_scaled <- scale(amplitudes_raw, center = TRUE, scale = FALSE)
# Create the parametric regressor
reg_amp <- regressor(onsets_amp, HRF_SPMG1, amplitude = amplitudes_scaled)
print(reg_amp)
# Plot the parametric regressor
scan_times_amp <- seq(0, max(onsets_amp) + 30, by = TR)
plot(reg_amp, grid = scan_times_amp)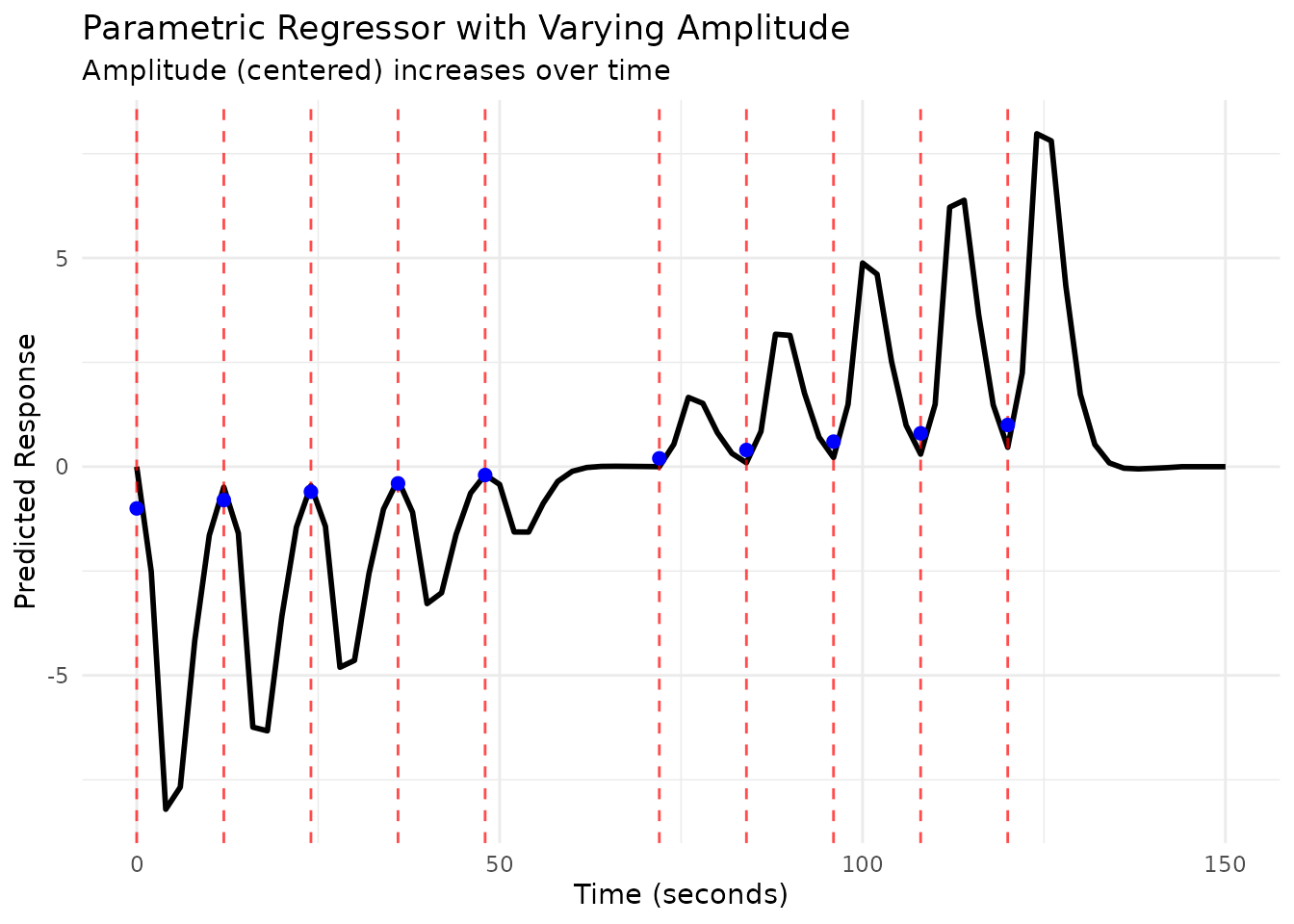
Combining Duration and Amplitude Modulation
You can provide both duration and amplitude
vectors to model events that vary in both aspects.
set.seed(123)
onsets_comb <- seq(0, 10 * 12, length.out = 11)
amps_comb <- scale(1:length(onsets_comb), center = TRUE, scale = FALSE)
durs_comb <- sample(1:5, length(onsets_comb), replace = TRUE)
reg_comb <- regressor(onsets_comb, HRF_SPMG1,
amplitude = amps_comb, duration = durs_comb)
print(reg_comb)
# Plot the combined regressor
scan_times_comb <- seq(0, max(onsets_comb) + 30, by = TR)
plot(reg_comb, grid = scan_times_comb)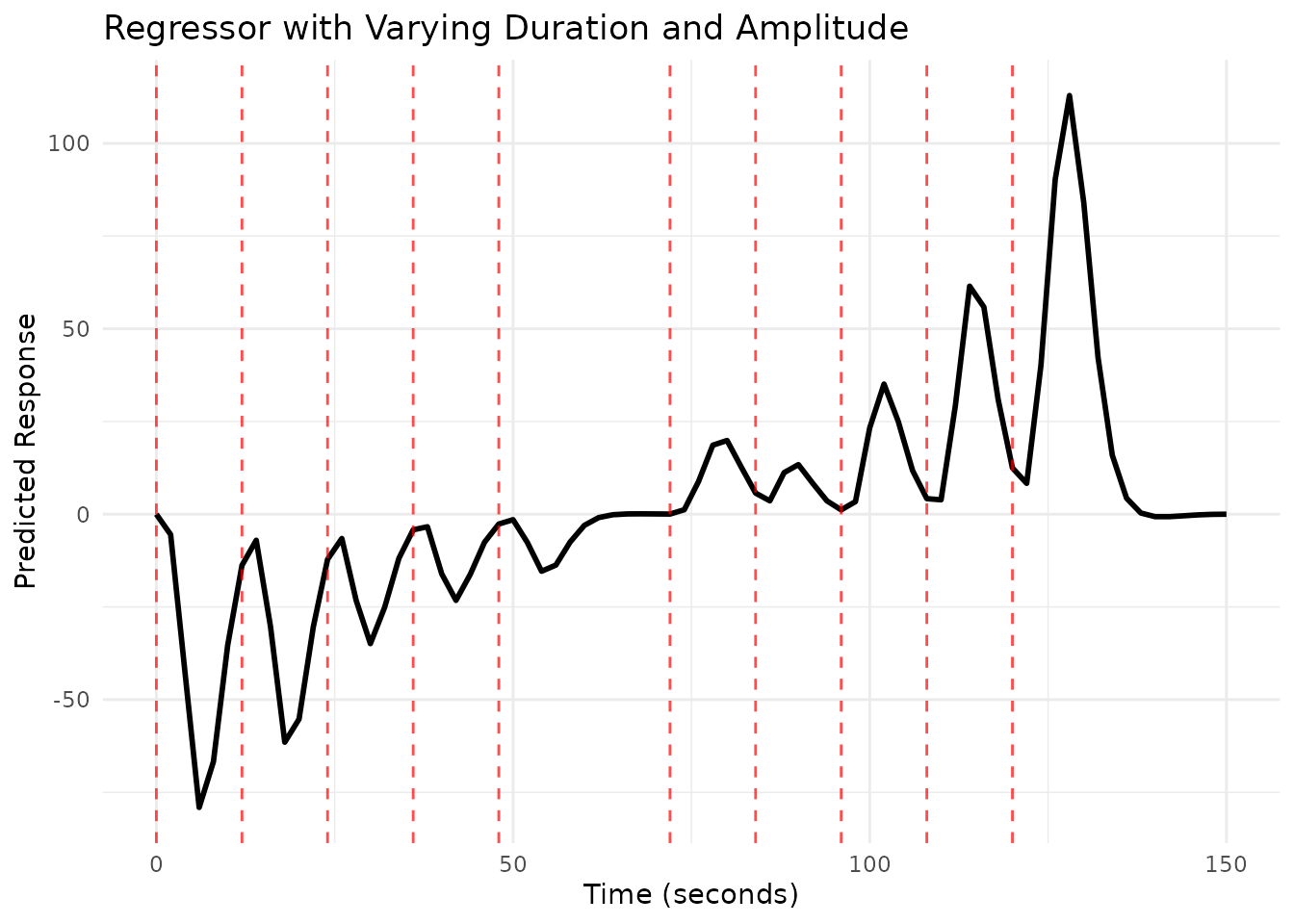
Regressors with HRF Basis Sets
If you use an HRF object with multiple basis functions (e.g.,
HRF_SPMG3, HRF_BSPLINE), the
regressor object will represent multiple timecourses, one
for each basis function. evaluate() will return a
matrix.
# Use a B-spline basis set
onsets_basis <- seq(0, 10 * 12, length.out = 11)
hrf_basis <- HRF_BSPLINE # Uses N=5 basis functions by default
reg_basis <- regressor(onsets_basis, hrf_basis)
print(reg_basis)
nbasis(reg_basis) # Should be 5
#> [1] 5
# Evaluate - this returns a matrix
scan_times_basis <- seq(0, max(onsets_basis) + 30, by = TR)
pred_basis_matrix <- evaluate(reg_basis, scan_times_basis)
dim(pred_basis_matrix) # rows = time points, cols = basis functions
#> [1] 76 5
# Plot using the plot() method - automatically handles multi-basis
plot(reg_basis, grid = scan_times_basis)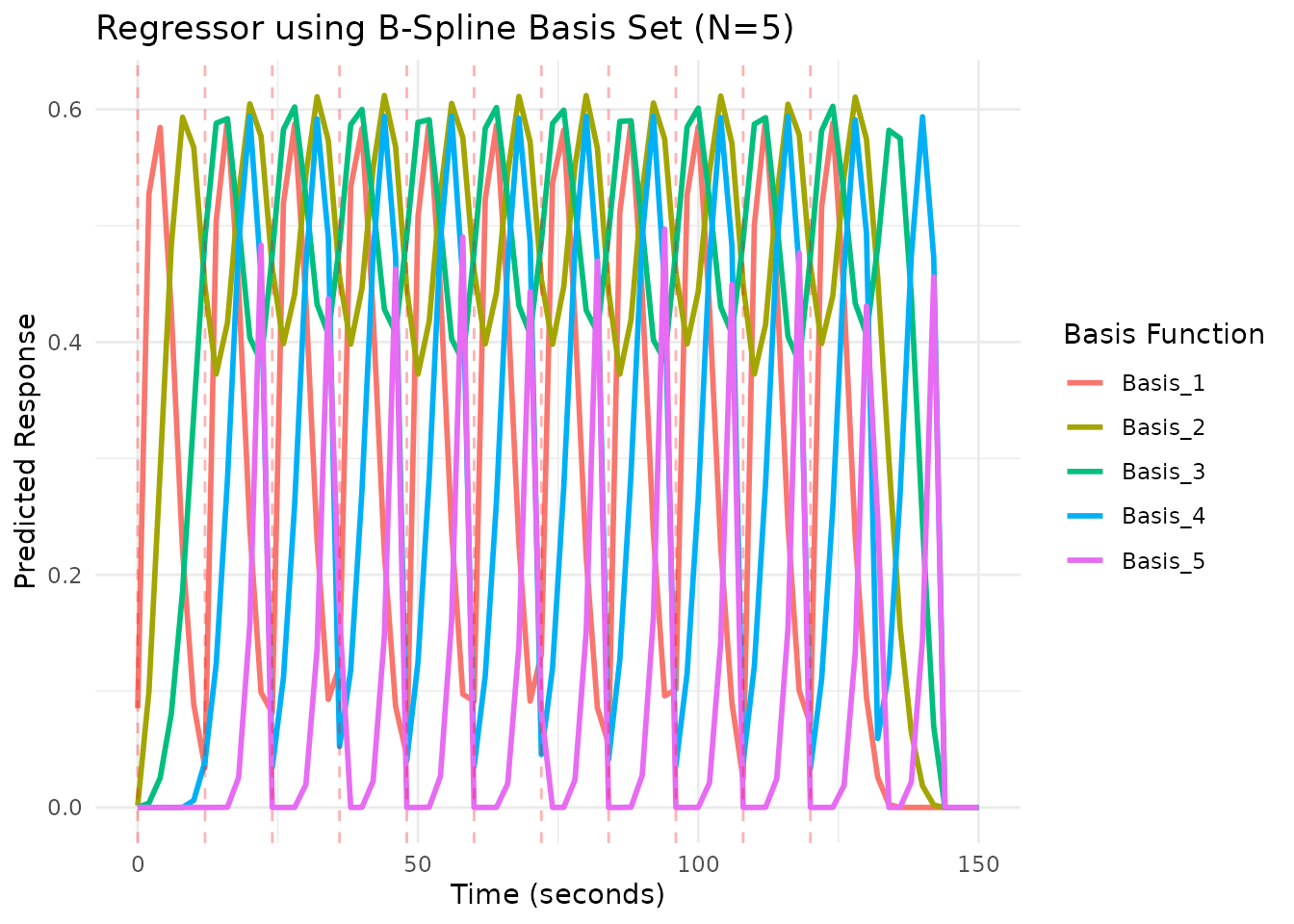
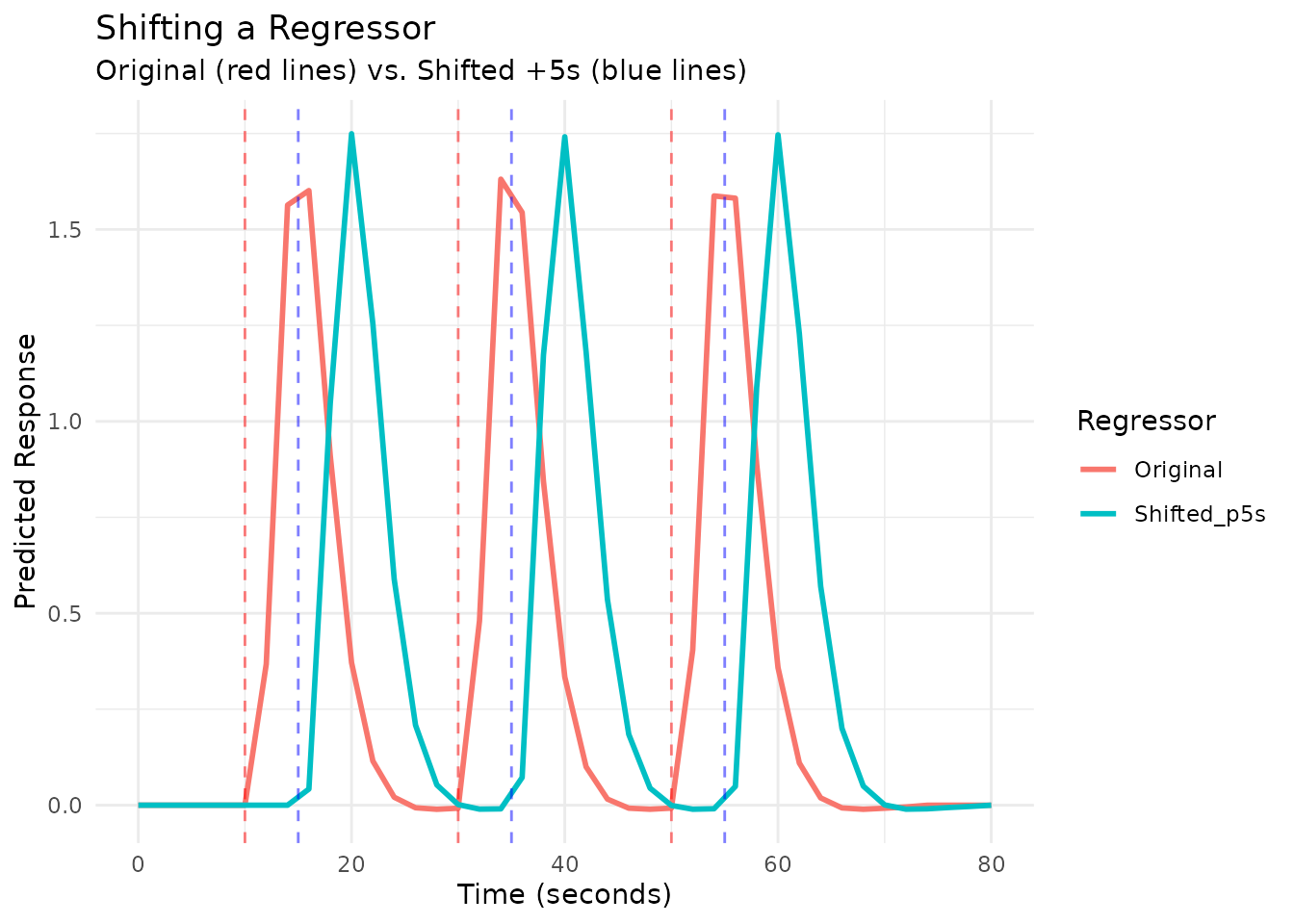
Shifting Regressors
You can temporally shift all onsets within a regressor using the
shift() method.
# Original regressor
reg_orig <- regressor(onsets = c(10, 30, 50), hrf = HRF_SPMG1)
# Shifted regressor (delay by 5 seconds)
reg_shifted <- shift(reg_orig, shift_amount = 5)
onsets(reg_orig)
#> [1] 10 30 50
onsets(reg_shifted) # Onsets are now 15, 35, 55
#> [1] 15 35 55
# Compare original and shifted using plot_regressors()
scan_times_shift <- seq(0, 80, by = TR)
plot_regressors(reg_orig, reg_shifted,
labels = c("Original", "Shifted +5s"),
grid = scan_times_shift,
show_onsets = TRUE, # Show onsets for both
title = "Shifting a Regressor")